6D6E
 
 | | Triclinic lysozyme (295 K) in the presence of 47% xylose | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D6F
 
 | |
6D6G
 
 | | Triclinic lysozyme (295 K) in the presence of 47% MPD | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D6H
 
 | | Triclinic lysozyme cryocooled to 100 K with 47% MPD as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Lysozyme C, NITRATE ION | | Authors: | Juers, D.H. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.000064 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D6I
 
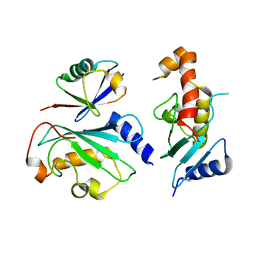 | | Ube2V1 in complex with ubiquitin variant Ubv.V1.1 and Ube2N/Ubc13 | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1, Ubv.V1.1 | | Authors: | Ceccarelli, D.F, Garg, P, Keszei, A, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-21 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D6J
 
 | |
6D6K
 
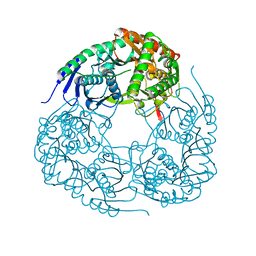 | |
6D6L
 
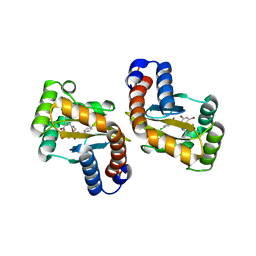 | |
6D6M
 
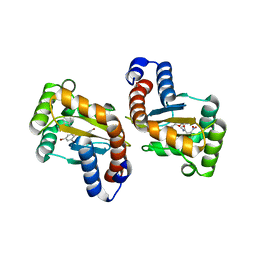 | |
6D6N
 
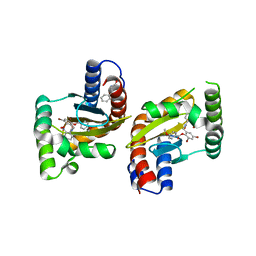 | | The structure of ligand binding domain of LasR in complex with TP-1 homolog, compound 16 | | Descriptor: | 2,4-dibromo-6-{[(2-nitrobenzene-1-carbonyl)amino]methyl}phenyl 4-methoxybenzoate, PHENYLALANINE, Transcriptional activator protein LasR | | Authors: | Dong, S.H, Nair, S.K. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Biochemical Studies of Non-native Agonists of the LasR Quorum-Sensing Receptor Reveal an L3 Loop "Out" Conformation for LasR.
Cell Chem Biol, 25, 2018
|
|
6D6O
 
 | |
6D6P
 
 | |
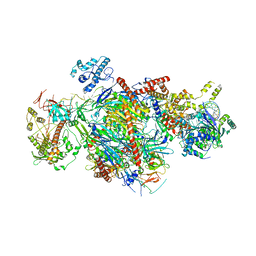
6D6Q
 
 | |
6D6R
 
 | |
6D6S
 
 | |
6D6T
 
 | | Human GABA-A receptor alpha1-beta2-gamma2 subtype in complex with GABA and flumazenil, conformation B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Noviello, C.M, Teng, J, Walsh Jr, R.M, Kim, J.J, Hibbs, R.E. | | Deposit date: | 2018-04-22 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structure of a human synaptic GABAAreceptor.
Nature, 559, 2018
|
|
6D6U
 
 | | Human GABA-A receptor alpha1-beta2-gamma2 subtype in complex with GABA and flumazenil, conformation A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Noviello, C.M, Teng, J, Walsh Jr, R.M, Kim, J.J, Hibbs, R.E. | | Deposit date: | 2018-04-22 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of a human synaptic GABAAreceptor.
Nature, 559, 2018
|
|
6D6V
 
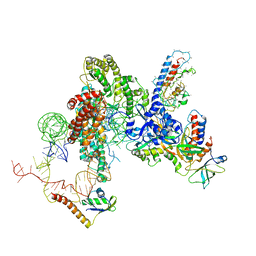 | | CryoEM structure of Tetrahymena telomerase with telomeric DNA at 4.8 Angstrom resolution | | Descriptor: | DNA (5'-D(P*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*GP*TP*TP*GP*GP*GP*G)-3'), RNA (159-MER), Telomerase associated protein p65, ... | | Authors: | Jiang, J, Wang, Y, Susac, L, Chan, H, Basu, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2018-04-22 | | Release date: | 2018-05-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of Telomerase with Telomeric DNA.
Cell, 173, 2018
|
|
6D6W
 
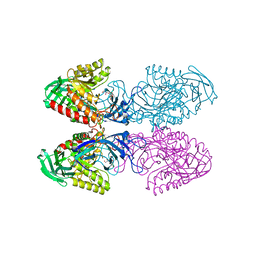 | | Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate | | Descriptor: | Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D6X
 
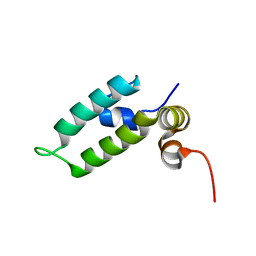 | | HSP40 co-chaperone Sis1 J-domain | | Descriptor: | Type II HSP40 co-chaperone | | Authors: | Pinheiro, G.M.S, Amorim, G.C, Iqbal, A, Ramos, C.H.I, Almeida, F.C.L. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR investigation on the structure and function of the isolated J-domain from Sis1: Evidence of transient inter-domain interactions in the full-length protein.
Arch.Biochem.Biophys., 669, 2019
|
|
6D6Y
 
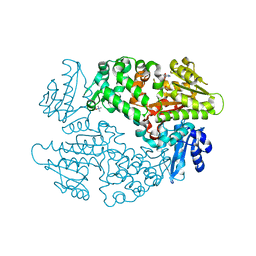 | |
6D6Z
 
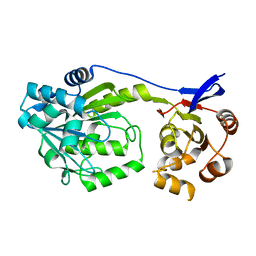 | |
6D71
 
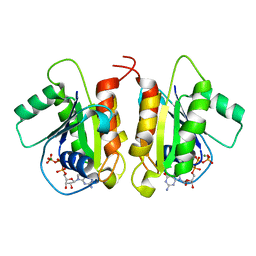 | | Crystal Structure of the Human Miro1 N-terminal GTPase bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 1 | | Authors: | Smith, K.P, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2018-04-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7180779 Å) | | Cite: | Insight into human Miro1/2 domain organization based on the structure of its N-terminal GTPase.
J.Struct.Biol., 212, 2020
|
|
6D72
 
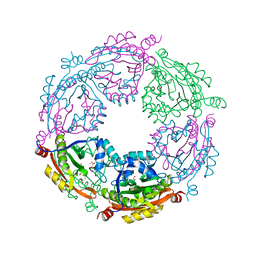 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions.
To Be Published
|
|
6D73
 
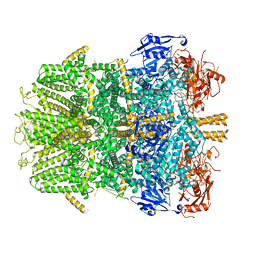 | | Cryo-EM structure of the zebrafish TRPM2 channel in the presence of Ca2+ | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel, subfamily M | | Authors: | Yin, Y, Wu, M, Borschel, W.F, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
