6D45
 
 | | L89S Mutant of FeBMb Sperm Whale Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhagi-Damodaran, A, Mirts, E.N, Sandoval, B, Lu, Y. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Heme redox potentials hold the key to reactivity differences between nitric oxide reductase and heme-copper oxidase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6D46
 
 | |
6D47
 
 | |
6D48
 
 | | Cell Surface Receptor | | Descriptor: | Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
6D49
 
 | | Cell Surface Receptor in Complex with Ligand at 1.80-A Resolution | | Descriptor: | 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, GLYCEROL, Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
6D4A
 
 | | Cell Surface Receptor with Bound Ligand at 1.75-A Resolution | | Descriptor: | 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, GLYCEROL, Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
6D4B
 
 | | Crystal structure of Candida boidinii formate dehydrogenase V123A mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
6D4C
 
 | | Crystal structure of Candida boidinii formate dehydrogenase V123G mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
6D4D
 
 | | Crystal structure of the PTP epsilon D1 domain | | Descriptor: | Receptor-type tyrosine-protein phosphatase epsilon | | Authors: | Lountos, G.T, Raran-Kurussi, S, Zhao, B.M, Dyas, B.K, Austin, B.P, Burke Jr, T.R, Ulrich, R.G, Waugh, D.S. | | Deposit date: | 2018-04-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | High-resolution crystal structures of the D1 and D2 domains of protein tyrosine phosphatase epsilon for structure-based drug design.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D4E
 
 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Fc Fragment of IgG1 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6D4F
 
 | | Crystal structure of PTP epsilon D2 domain (A455N/V457Y/E597D) | | Descriptor: | PENTAETHYLENE GLYCOL, Receptor-type tyrosine-protein phosphatase epsilon | | Authors: | Lountos, G.T, Raran-Kurussi, S, Zhao, B.M, Dyas, B.K, Austin, B.P, Burke Jr, T.R, Ulrich, R.G, Waugh, D.S. | | Deposit date: | 2018-04-18 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | High-resolution crystal structures of the D1 and D2 domains of protein tyrosine phosphatase epsilon for structure-based drug design.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6D4G
 
 | | N-GTPase domain of p190RhoGAP-A | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rho GTPase-activating protein 35 | | Authors: | Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2018-04-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The N-Terminal GTPase Domain of p190RhoGAP Proteins Is a PseudoGTPase.
Structure, 26, 2018
|
|
6D4I
 
 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG2 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6D4K
 
 | |
6D4L
 
 | |
6D4M
 
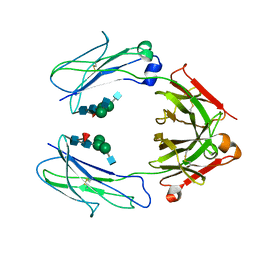 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG3 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6D4N
 
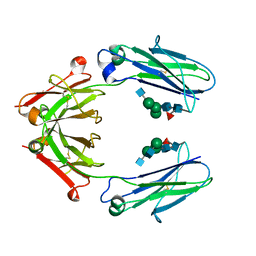 | | Crystal structure of a Fc fragment of Rhesus macaque (Macaca mulatta) IgG4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG4 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6D4O
 
 | | Eubacterium eligens beta-glucuronidase bound to an amoxapine-glucuronide conjugate | | Descriptor: | (5aR,9aR)-2-chloro-11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5a,6,9,9a-tetrahydrodibenzo[b,f][1,4]oxazepine, Beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Pellock, S.J, Walton, W.G, Redinbo, M.R. | | Deposit date: | 2018-04-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut Microbial beta-Glucuronidase Inhibition via Catalytic Cycle Interception.
ACS Cent Sci, 4, 2018
|
|
6D4P
 
 | | Ube2D1 in complex with ubiquitin variant Ubv.D1.1 | | Descriptor: | Ubiquitin Variant Ubv.D1.1, Ubiquitin-conjugating enzyme E2 D1 | | Authors: | Ceccarelli, D.F, Garg, P, Sidhu, S, Sicheri, F. | | Deposit date: | 2018-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and Functional Analysis of Ubiquitin-based Inhibitors That Target the Backsides of E2 Enzymes.
J.Mol.Biol., 432, 2020
|
|
6D4Q
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 14 (VCC900455) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, cycloheptyl{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}methanone | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4R
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 18 (VCC399134) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, hydroxy(3-{4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carbonyl}phenyl)oxoammonium | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4S
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 37 (VCC670597) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, N-(2,3-dichlorophenyl)-4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carboxamide | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4T
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 45 (VCC117054) | | Descriptor: | (2S)-N-(2H-1,3-benzodioxol-5-yl)-4-[(isoquinolin-5-yl)sulfonyl]-2-methylpiperazine-1-carboxamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4U
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 27 (VCC663664) | | Descriptor: | 2-(2,4-dimethoxyphenyl)-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D4V
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 22 (VCC061422) | | Descriptor: | 2-cyclohexyl-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
