5CCF
 
 | |
8RVC
 
 | | Crystal structure of alpha keto acid C-methyl-transferases MrsA bound to ketoarginine | | Descriptor: | 1,2-ETHANEDIOL, 2-ketoarginine methyltransferase, 5-[(diaminomethylidene)amino]-2-oxopentanoic acid, ... | | Authors: | Gerhardt, S, Kemper, F, Andexer, J.N. | | Deposit date: | 2024-02-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structures and Protein Engineering of the alpha-Keto Acid C-Methyltransferases SgvM and MrsA for Rational Substrate Transfer.
Chembiochem, 25, 2024
|
|
7YY0
 
 | |
7PFG
 
 | |
7N89
 
 | |
8V3N
 
 | | CCP5 in complex with Glu-P-Glu transition state analog | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
7Z70
 
 | |
6MTV
 
 | | Crystal structure of human Scribble PDZ1:MCC complex | | Descriptor: | Colorectal mutant cancer protein, DI(HYDROXYETHYL)ETHER, Protein scribble homolog, ... | | Authors: | Caria, S, Stewart, B.Z, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural analysis of phosphorylation-associated interactions of human MCC with Scribble PDZ domains.
Febs J., 286, 2019
|
|
7PF7
 
 | | Apo structure of SynFtn variant D65A | | Descriptor: | CHLORIDE ION, Ferritin, SODIUM ION | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
5CDC
 
 | |
8UAT
 
 | | Thermus scotoductus SA-01 Ene-reductase Compound 3b Complex | | Descriptor: | 1-[2-(4-hydroxyphenyl)ethyl]-1,4-dihydropyridine-3-carboxamide, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
7YY5
 
 | |
5CDL
 
 | | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Proline dipeptidase | | Authors: | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R. | | Deposit date: | 2015-07-04 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proline dipeptidase from Deinococcus radiodurans (selenomethionine derivative)
To Be Published
|
|
7VNS
 
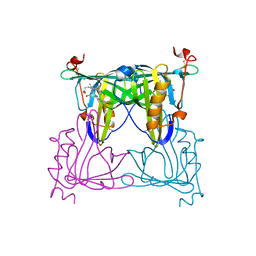 | | Sandercyanin mutant E79A-Biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Yadav, K, Ghosh, S, Subramanian, R. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phenylalanine stacking enhances the red fluorescence of biliverdin IX alpha on UV excitation in sandercyanin fluorescent protein.
Febs Lett., 596, 2022
|
|
8RUN
 
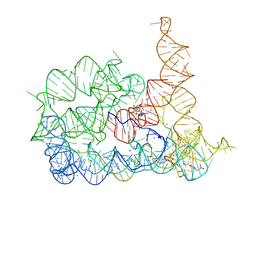 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+, and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Domains 1-5, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
8V3O
 
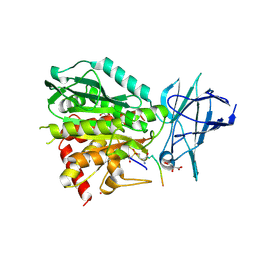 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8OFR
 
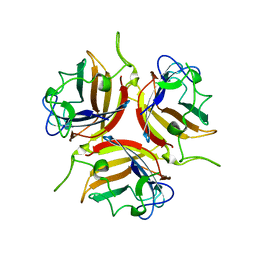 | | Human adenovirus type 25 fiber-knob protein complexed with sialic acid | | Descriptor: | Fiber, N-acetyl-beta-neuraminic acid | | Authors: | Rizkallah, P.J, Parker, A.L, Mundy, R.M, Baker, A.T. | | Deposit date: | 2023-03-16 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus.
Npj Viruses, 1, 2023
|
|
7NLS
 
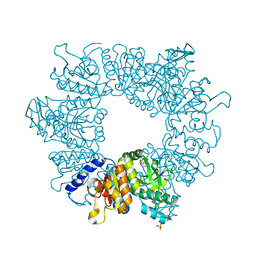 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with methyl 4-hydroxy-3-iodobenzoate | | Descriptor: | Acetylglutamate kinase, SULFATE ION, methyl 3-iodanyl-4-oxidanyl-benzoate | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8OFQ
 
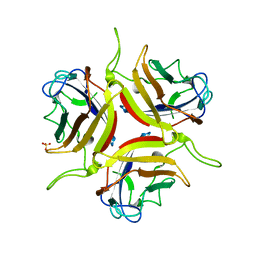 | | Human adenovirus type 25 fiber-knob protein | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fiber, ... | | Authors: | Rizkallah, P.J, Parker, A.L, Mundy, R.M, Baker, A.T. | | Deposit date: | 2023-03-16 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus.
Npj Viruses, 1, 2023
|
|
8R4Z
 
 | |
8SC7
 
 | |
7YY1
 
 | |
7NM7
 
 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM | | Descriptor: | Antimycin pathway standalone ketoreductase enzyme, AntM | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
7YY9
 
 | |
8V3M
 
 | | CCP5 apo structure | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
