6N6S
 
 | | Crystal structure of ABIN-1 UBAN | | Descriptor: | TNFAIP3-interacting protein 1 | | Authors: | Rahighi, S, Dikic, I, Wakatsuki, S. | | Deposit date: | 2018-11-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
J.Mol.Biol., 431, 2019
|
|
3MXM
 
 | |
3RM7
 
 | | CDK2 in complex with inhibitor KVR-1-91 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4-hydroxybenzyl)amino]-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R5I
 
 | | Crystal structure of liganded Hemoglobin complexed with a potent Antisickling agent, INN-312 | | Descriptor: | 5-methoxy-2-(pyridin-3-ylmethoxy)benzaldehyde, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Safo, M.K, Musayev, F.N, Safo, R.P, Daniels, D, Eseonu, D.N, Parra, J. | | Deposit date: | 2011-03-18 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and in Vitro Chracterization of Pyridyl Derivatives of Benzaldehydes : Highly Potent Antisickling Agents
To be Published
|
|
6MQH
 
 | |
3RAL
 
 | | CDK2 in complex with inhibitor RC-2-34 | | Descriptor: | 4-{[4-amino-5-(3-methoxybenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3R1S
 
 | | CDK2 in complex with inhibitor KVR-1-127 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(6-chloropyridin-3-yl)methyl]amino}-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-11 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3OKK
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
6MWI
 
 | |
6MSS
 
 | | Diversity in the type II Natural Killer T cell receptor repertoire and antigen specificity leads to differing CD1d docking strategies | | Descriptor: | (2S)-2-(heptadecanoyloxy)-3-{[(10S)-10-methyloctadecanoyl]oxy}propyl alpha-D-glucopyranosiduronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sundararaj, S, Le Nours, J, Praveena, T, Rossjohn, J. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Distinct CD1d docking strategies exhibited by diverse Type II NKT cell receptors.
Nat Commun, 10, 2019
|
|
3R6B
 
 | | Crystal Structure of Thrombospondin-1 TSR Domains 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1 | | Authors: | Page, R.C, Klenotic, P.A, Misra, S, Silverstein, R.L. | | Deposit date: | 2011-03-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, purification and structural characterization of functionally replete thrombospondin-1 type 1 repeats in a bacterial expression system.
Protein Expr.Purif., 80, 2011
|
|
6MX0
 
 | | Crystal structure of human STING apoprotein (G230A, H232R, R293Q) | | Descriptor: | CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A, Siu, T, Ho, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING.
ACS Med Chem Lett, 10, 2019
|
|
3RAH
 
 | | CDK2 in complex with inhibitor RC-2-22 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(cyclohexylamino)-1,3-thiazol-5-yl](naphthalen-2-yl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
6MZW
 
 | | TAS-120 covalent complex with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
3RPG
 
 | | Bmi1/Ring1b-UbcH5c complex structure | | Descriptor: | E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | Bentley, M.L, Dong, K.C, Cochran, A.G. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6485 Å) | | Cite: | Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex.
Embo J., 30, 2011
|
|
6NMP
 
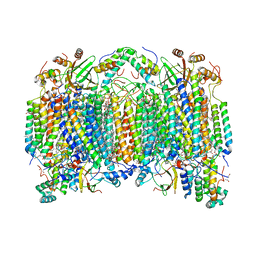 | | SFX structure of oxidized cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3R1Q
 
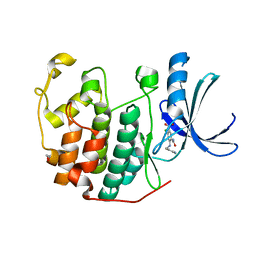 | | CDK2 in complex with inhibitor KVR-1-102 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-{[(6-chloropyridin-3-yl)methyl]amino}-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-11 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
6NQT
 
 | | GalNac-T2 soaked with UDP-sugar | | Descriptor: | MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{R})-3-(hex-5-ynoylamino)-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Fernandez, D, Bertozzi, C.R, Schumann, B, Agbay, A. | | Deposit date: | 2019-01-21 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
3R73
 
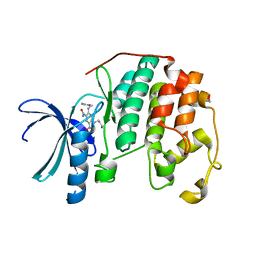 | | CDK2 in complex with inhibitor KVR-1-164 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-aminopropyl)amino]-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3MWR
 
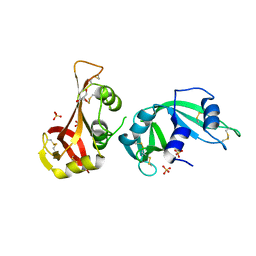 | |
3R8V
 
 | | CDK2 in complex with inhibitor RC-1-135 | | Descriptor: | Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](3-nitrophenyl)methanone | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
6NMF
 
 | | SFX structure of reduced cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-10 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3RA5
 
 | | Crystal structure of T. celer L30e E6A/R92A variant | | Descriptor: | 50S ribosomal protein L30e, SULFATE ION | | Authors: | Chan, C.H, Wong, K.B. | | Deposit date: | 2011-03-27 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing salt-bridge enhances protein thermostability by reducing the heat capacity change of unfolding
Plos One, 6, 2011
|
|
3RFN
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_1wnu_001, ZINC ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
6LM1
 
 | | The crystal structure of cyanorhodopsin (CyR) N4075R from cyanobacteria Tolypothrix sp. NIES-4075 | | Descriptor: | DECANE, DODECANE, HEXADECANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2019-12-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
