3JRG
 
 | |
3JRQ
 
 | | Crystal structure of (+)-ABA-bound PYL1 in complex with ABI1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Protein phosphatase 2C 56, Putative uncharacterized protein At5g46790 | | Authors: | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | Deposit date: | 2009-09-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
3JS4
 
 | |
3JSQ
 
 | | Crystal structure of adipocyte fatty acid binding protein non-covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, Adipocyte fatty acid-binding protein, CHLORIDE ION, ... | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
3JTW
 
 | |
3JUK
 
 | |
3JYG
 
 | |
3JSP
 
 | |
3JTY
 
 | | Crystal structure of a BenF-like porin from Pseudomonas fluorescens Pf-5 | | Descriptor: | BenF-like porin, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Sampathkumar, P, Lu, F, Zhao, X, Wasserman, S, Iuzuka, M, Bain, K, Rutter, M, Gheyi, T, Atwell, S, Luz, J, Gilmore, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Proteins, 78, 2010
|
|
3JV7
 
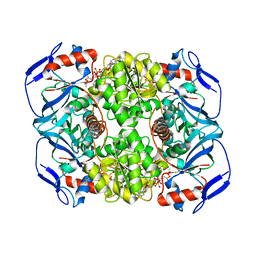 | | Structure of ADH-A from Rhodococcus ruber | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ADH-A, ... | | Authors: | Karabec, M, Lyskowski, A, Gruber, K. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into substrate specificity and solvent tolerance in alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541.
Chem.Commun.(Camb.), 2010
|
|
3JWG
 
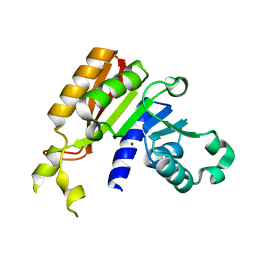 | | Crystal structure analysis of the methyltransferase domain of bacterial-CtHen1-C | | Descriptor: | MAGNESIUM ION, Methyltransferase type 12 | | Authors: | Huang, R.H, Chan, C.M, Zhou, C, Brunzelle, J.S. | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical insights into 2'-O-methylation at the 3'-terminal nucleotide of RNA by Hen1.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3JWO
 
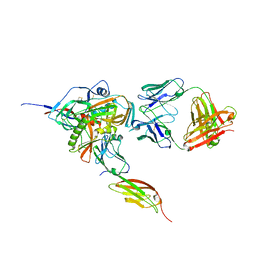 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JX9
 
 | |
3JXH
 
 | | CA-like domain of human PTPRG | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, SULFATE ION | | Authors: | Bouyain, S. | | Deposit date: | 2009-09-19 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JZD
 
 | |
3K0D
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3JPZ
 
 | | Crystal Structure of Lombricine Kinase | | Descriptor: | Lombricine kinase, NITRATE ION | | Authors: | Bush, D.J, Kirillova, O, Clark, S.A, Fabiola, F, Somasundaram, T, Chapman, M.S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of lombricine kinase: implications for phosphagen kinase conformational changes.
J.Biol.Chem., 286, 2011
|
|
3K12
 
 | | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, uncharacterized protein A6V7T0 | | Authors: | Ho, M, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa
To be published
|
|
3JQD
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor 2-amino-4-oxo-6-phenyl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile (DX7) | | Descriptor: | 2-amino-4-oxo-6-phenyl-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2009-09-06 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
3K1P
 
 | |
3JRC
 
 | |
3JRI
 
 | |
3JS9
 
 | |
3K2J
 
 | | Crystal Structure of the 3rd Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) | | Descriptor: | CHLORIDE ION, Protein polybromo-1, SULFATE ION | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Chaikuad, A, Pike, A.C.W, Krojer, T, Sethi, R, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the 3rd Bromodomain of Human Poly-bromodomain containing protein 1 (PB1)
To be Published
|
|
3JSG
 
 | |
