1ESL
 
 | |
3E1Q
 
 | | Crystal structure of W133F variant E. coli Bacterioferritn with iron. | | Descriptor: | BACTERIOFERRITIN, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crow, A, Lawson, T.L, Lewin, A, Moore, G.R, Le Brun, N.E. | | Deposit date: | 2008-08-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Monitoring the iron status of the ferroxidase center of Escherichia coli bacterioferritin using fluorescence spectroscopy.
Biochemistry, 48, 2009
|
|
8VBB
 
 | |
8UXQ
 
 | |
7ZI0
 
 | | Structure of human Smoothened in complex with cholesterol and SAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-chloro-N-[trans-4-(methylamino)cyclohexyl]-N-{[3-(pyridin-4-yl)phenyl]methyl}-1-benzothiophene-2-carboxamide, CHOLESTEROL, ... | | Authors: | Byrne, E.F.X, Woolley, R.E, Ansell, B, Sansom, M.S.P, Newstead, S, Siebold, C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Patched 1 regulates Smoothened by controlling sterol binding to its extracellular cysteine-rich domain.
Sci Adv, 8, 2022
|
|
5F6C
 
 | |
1S1X
 
 | | Crystal structure of V108I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
7ZAI
 
 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA, mRNA and aIF1A. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
6O5R
 
 | |
8XXV
 
 | | Cryo-EM Structure of the Prostaglandin D2 Receptor 2-indomethacin Coupled to G Protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-01-19 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of lipid and ligand regulation of prostaglandin receptor DP2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XXU
 
 | | Cryo-EM Structure of the Prostaglandin D2 Receptor 2 Coupled to G Protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-01-19 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Molecular basis of lipid and ligand regulation of prostaglandin receptor DP2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8503 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6O5S
 
 | |
3MEG
 
 | | HIV-1 K103N Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
7ZHG
 
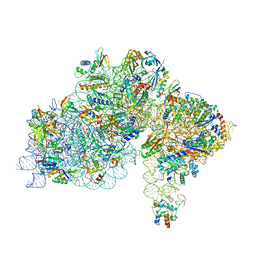 | | High-resolution cryo-EM structure of Pyrococcus abyssi 30S ribosomal subunit bound to mRNA and initiator tRNA anticodon stem-loop | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Kazan, R, Bourgeois, G, Mechulam, Y, Coureux, P.D, Schmitt, E. | | Deposit date: | 2022-04-06 | | Release date: | 2022-06-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
4Z3N
 
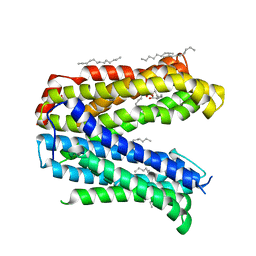 | | Crystal structure of the MATE transporter ClbM | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CACODYLATE ION, Putative drug/sodium antiporter | | Authors: | Mousa, J.J, Bruner, S.D. | | Deposit date: | 2015-03-31 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | MATE transport of the E. coli-derived genotoxin colibactin.
Nat Microbiol, 1, 2016
|
|
7RB7
 
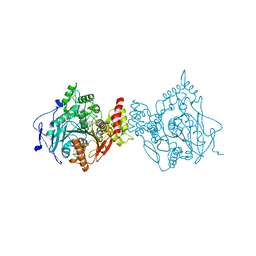 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA and MMB4 oxime | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
8W6G
 
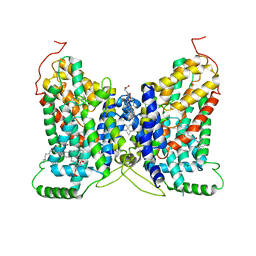 | | NaDC1 with inhibitor ACA | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-[[(~{E})-3-(4-pentylphenyl)prop-2-enoyl]amino]benzoic acid, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chi, X, Chen, Y, Li, Y, Zhang, Y, Shen, Y, Chen, Y, Wang, Z, Yan, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human NaS1 and NaDC1 transporters revealed the elevator transport and allosteric regulation mechanism.
Sci Adv, 10, 2024
|
|
4YJ2
 
 | | Crystal structure of tubulin bound to MI-181 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,6-dimethyl-2-[(E)-2-(pyridin-3-yl)ethenyl]-1,3-benzothiazole, CALCIUM ION, ... | | Authors: | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
6ND5
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with chloramphenicol and bound to mRNA and A-, P-, and E-site tRNAs at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
3IMQ
 
 | | Crystal structure of the NusB101-S10(delta loop) complex | | Descriptor: | 30S ribosomal protein S10, N utilization substance protein B, POTASSIUM ION | | Authors: | Luo, X, Wahl, M.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination.
Nucleic Acids Res., 38, 2010
|
|
1D7A
 
 | | CRYSTAL STRUCTURE OF E. COLI PURE-MONONUCLEOTIDE COMPLEX. | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, PHOSPHORIBOSYLAMINOIMIDAZOLE CARBOXYLASE | | Authors: | Mathews, I.I, Kappock, T.J, Stubbe, J, Ealick, S.E. | | Deposit date: | 1999-10-16 | | Release date: | 1999-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway.
Structure Fold.Des., 7, 1999
|
|
6ND6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with erythromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.85A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-02-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
9C12
 
 | | Leukotriene C4-bound Multidrug Resistance-associated Protein 2 (MRP2) | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, ATP-binding cassette sub-family C member 2, CHOLESTEROL, ... | | Authors: | Koide, E, Chen, J. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis for the transport and regulation mechanism of the multidrug resistance-associated protein 2.
Nat Commun, 16, 2025
|
|
8HAQ
 
 | | The complex of Src with GW8510 | | Descriptor: | 4-[(~{E})-(7-oxidanyl-6~{H}-pyrrolo[2,3-g][1,3]benzothiazol-8-yl)methylideneamino]-~{N}-pyridin-2-yl-benzenesulfonamide, Isoform 2 of Proto-oncogene tyrosine-protein kinase Src | | Authors: | Zhu, S.J, Bi, S.Z. | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The complex of Src with GW8510
To Be Published
|
|
