7BJ3
 
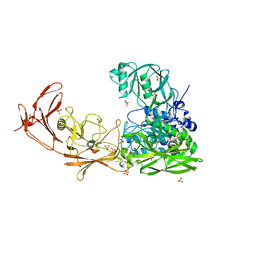 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
7BIJ
 
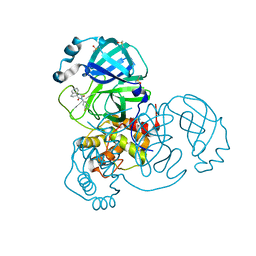 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 13 | | Descriptor: | (3~{S})-3'-(5-fluoranylpyridin-3-yl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7KXJ
 
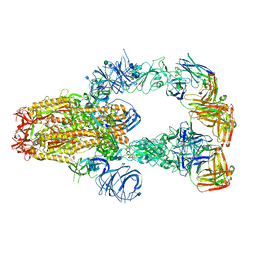 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 3-"up", asymmetric | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KJ4
 
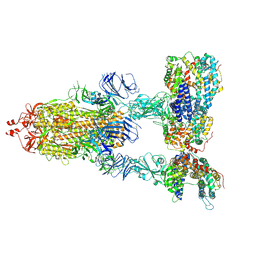 | | SARS-CoV-2 Spike Glycoprotein with three ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KLG
 
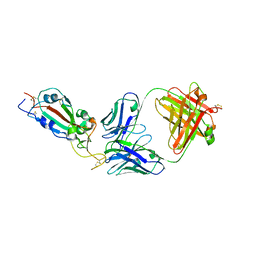 | | SARS-CoV-2 RBD in complex with Fab 15033 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033 heavy chain, Fab 15033 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KML
 
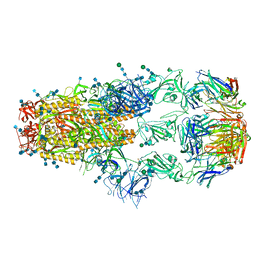 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, three RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KLH
 
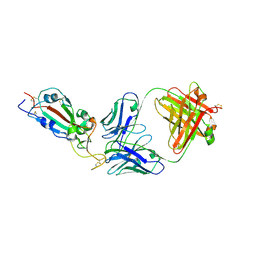 | | SARS-CoV-2 RBD in complex with Fab 15033-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, Fab 15033-7 light chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KMK
 
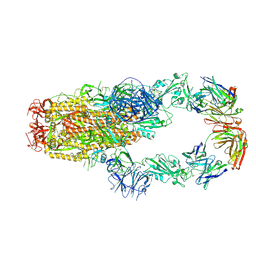 | | cryo-EM structure of SARS-CoV-2 spike in complex with Fab 15033-7, two RBDs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7L12
 
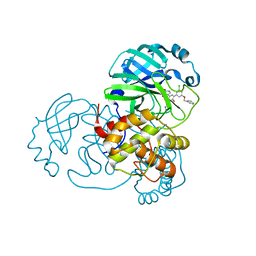 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 14 | | Descriptor: | (5S)-5-{3-[3-(benzyloxy)-5-chlorophenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L14
 
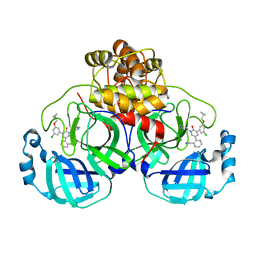 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 26 | | Descriptor: | 2-{3-[3-chloro-5-(cyclopropylmethoxy)phenyl]-2-oxo[2H-[1,3'-bipyridine]]-5-yl}benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L13
 
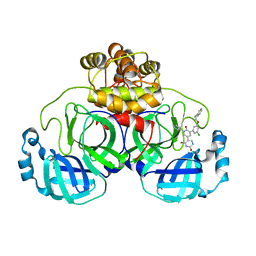 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5S)-5-(3-{3-chloro-5-[(2-chlorophenyl)methoxy]phenyl}-2-oxo[2H-[1,3'-bipyridine]]-5-yl)pyrimidine-2,4(3H,5H)-dione, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L10
 
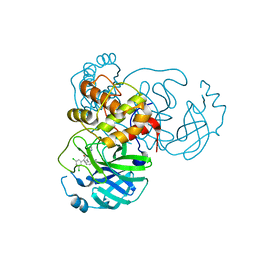 | | CRYSTAL STRUCTURE OF THE SARS-COV-2 (2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 2-[3-(3,5-dichlorophenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-13 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7L11
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 2-[3-(3-chloro-5-propoxyphenyl)-2-oxo[2H-[1,3'-bipyridine]]-5-yl]benzonitrile, 3C-like proteinase | | Authors: | Deshmukh, M.G, Ippolito, J.A, Stone, E.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Acs Cent.Sci., 7, 2021
|
|
7JJC
 
 | |
7KJ3
 
 | | SARS-CoV-2 Spike Glycoprotein with two ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KXK
 
 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 2-"up"-1-"down" conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
7KJ5
 
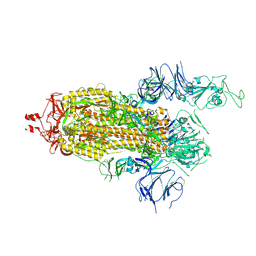 | | SARS-CoV-2 Spike Glycoprotein, prefusion with one RBD up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KJ2
 
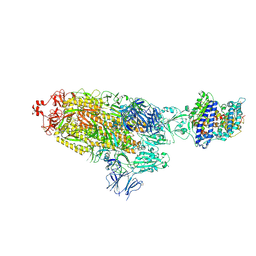 | | SARS-CoV-2 Spike Glycoprotein with one ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Chen, B. | | Deposit date: | 2020-10-25 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7TZ0
 
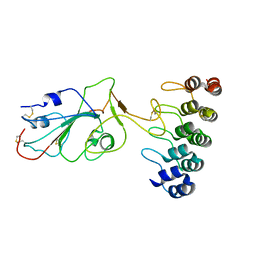 | |
7TYZ
 
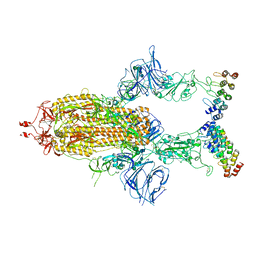 | | Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DARPin FSR22, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
7U0Q
 
 | |
7U0X
 
 | |
7MLF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C7 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
7MLG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C63 | | Descriptor: | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide, 3C-like proteinase | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
7UW3
 
 | |
