2KNR
 
 | | Solution structure of protein Atu0922 from A. tumefaciens. Northeast Structural Genomics Consortium target AtT13. Ontario Center for Structural Proteomics target ATC0905 | | Descriptor: | Uncharacterized protein ATC0905 | | Authors: | Gutmanas, A, Yee, S, Lemak, A, Fares, A, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein atc0905 from A. tumefaciens
To be Published
|
|
3M6M
 
 | | Crystal structure of RpfF complexed with REC domain of RpfC | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Cheng, Z, Lim, S.C, Qamra, R, Song, H. | | Deposit date: | 2010-03-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Sensor-Synthase Interaction in Autoinduction of the Quorum Sensing Signal DSF Biosynthesis
Structure, 18, 2010
|
|
2PQ3
 
 | | N-Terminal Calmodulin Zn-Trapped Intermediate | | Descriptor: | CACODYLATE ION, Calmodulin, ZINC ION | | Authors: | Warren, J.T, Guo, Q, Tang, W.J. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A 1.3-A structure of zinc-bound N-terminal domain of calmodulin elucidates potential early ion-binding step.
J.Mol.Biol., 374, 2007
|
|
2KFE
 
 | | Solution structure of meucin-24 | | Descriptor: | meucin-24 | | Authors: | Du, W, Xu, J, Gao, B, Zhu, S. | | Deposit date: | 2009-02-18 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of two linear cationic antimalarial peptides in the scorpion Mesobuthus eupeus
Biochimie, 92, 2010
|
|
2K9B
 
 | | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy | | Descriptor: | Dermadistinctin-K | | Authors: | Moraes, C.M, Verly, R.M, Resende, J.M, Aisenbrey, C, Bemquerer, M.P, Pilo-Veloso, D, Valente, A, Almeida, F.C.L, Bechinger, B. | | Deposit date: | 2008-10-07 | | Release date: | 2009-04-14 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy.
Biophys.J., 96, 2009
|
|
6DJK
 
 | |
2X8N
 
 | | Solution NMR structure of uncharacterized protein CV0863 from Chromobacterium violaceum. NORTHEAST STRUCTURAL GENOMICS TARGET (NESG) target CvT3. OCSP target CV0863. | | Descriptor: | CV0863 | | Authors: | Gutmanas, A, Fares, C, Yee, A, Lemak, A, Semesi, A, Arrowsmith, C.H, Ontario Centre for Structural Proteomics (OCSP), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Uncharacterized Protein Cv0863 from Chromobacterium Violaceum
To be Published
|
|
2GV1
 
 | |
2HS4
 
 | | T. maritima PurL complexed with FGAR and AMPPCP | | Descriptor: | MAGNESIUM ION, N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, ... | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
2XGJ
 
 | | Structure of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT RNA HELICASE DOB1, RNA (5'-(*AP*AP*AP*AP*A)-3') | | Authors: | Weir, J.R, Bonneau, F, Hentschel, J, Conti, E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis Reveals the Characteristic Features of Mtr4, a Dexh Helicase Involved in Nuclear RNA Processing and Surveillance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2NPV
 
 | |
2MTW
 
 | | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit into HLA-DR 1*03 molecules induces antibody production and protection | | Descriptor: | Erythrocyte-binding antigen 175 | | Authors: | Cifuentes, G, Salazar, L, Vargas, L, Parra, C, Vanegas, M, Cortes, J, Sandoval, M, Patarroyo, M.E. | | Deposit date: | 2014-09-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evidence supporting the hypothesis that specifically modifying a malaria peptide to fit HLA-DR 1*03 molecules induces antibody production and protection
To be Published
|
|
2MAX
 
 | |
4X4R
 
 | |
6I2T
 
 | | CryoEM reconstruction of full-length, fully-glycosylated human butyrylcholinesterase tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, lamellipodin-derived polyproline peptide | | Authors: | Leung, M.R, van Bezouwen, L.S, Schopfer, L.M, Sussman, J.L, Silman, I, Lockridge, O, Zeev-Ben-Mordehai, T. | | Deposit date: | 2018-11-01 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structure of the native butyrylcholinesterase tetramer reveals a dimer of dimers stabilized by a superhelical assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4XHN
 
 | | Bacillus thuringiensis ParM with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2015-01-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
2RND
 
 | |
4XF7
 
 | | myo-inositol 3-kinase bound with its substrates (AMPPCP and myo-inositol) | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Carbohydrate/pyrimidine kinase, PfkB family, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure and Product Analysis of an Archaeal myo-Inositol Kinase Reveal Substrate Recognition Mode and 3-OH Phosphorylation
Biochemistry, 54, 2015
|
|
7Z8N
 
 | | GacS histidine kinase from Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, Histidine kinase, R-1,2-PROPANEDIOL | | Authors: | Fadel, F, Bassim, V, Francis, V.I, Porter, S.L, Botzanowski, T, Legrand, P, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-03-17 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
1SAC
 
 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
6J6I
 
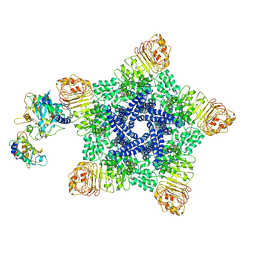 | | Reconstitution and structure of a plant NLR resistosome conferring immunity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-15 | | Release date: | 2019-03-20 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Reconstitution and structure of a plant NLR resistosome conferring immunity.
Science, 364, 2019
|
|
6J5T
 
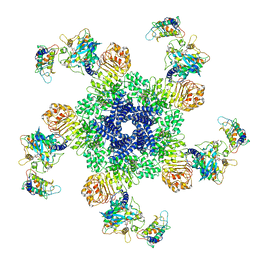 | | Reconstitution and structure of a plant NLR resistosome conferring immunity | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Reconstitution and structure of a plant NLR resistosome conferring immunity.
Science, 364, 2019
|
|
6JCS
 
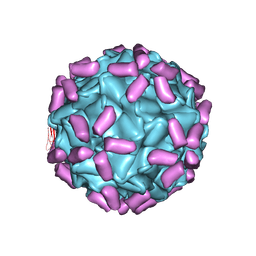 | | AAV5 in complex with AAVR | | Descriptor: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
2RMY
 
 | |
6JCR
 
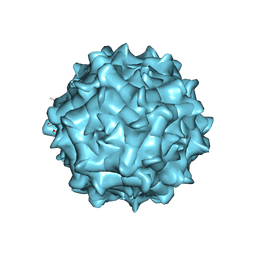 | | AAV1 in neutral condition at 3.07 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
