7PVU
 
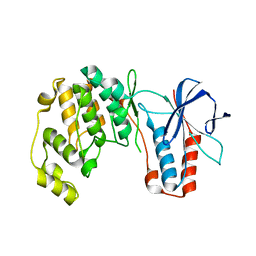 | | Crystal structure of p38alpha C162S in complex with CAS2094511-69-8, P 1 21 1 | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)-2-fluorobenzene-1-sulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2021-10-05 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
7O86
 
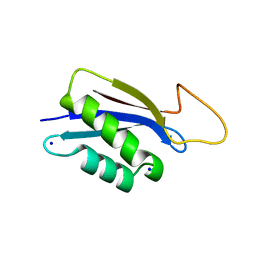 | | 1.73A X-ray crystal structure of the conserved C-terminal (CCT) of human SPAK | | Descriptor: | CALCIUM ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Elvers, K.T, Bax, B.D, Lipka-Lloyd, M, Mehellou, Y. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structures of the Human SPAK and OSR1 Conserved C-Terminal (CCT) Domains.
Chembiochem, 23, 2022
|
|
7AMN
 
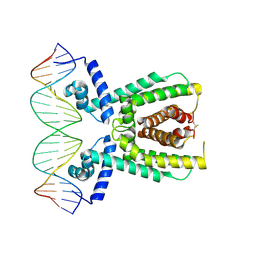 | | Structure of LuxR with DNA (repression) | | Descriptor: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
8PEK
 
 | |
6V1M
 
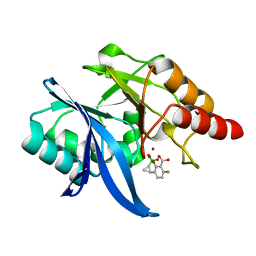 | | Structure of NDM-1 bound to QPX7728 at 1.05 A | | Descriptor: | (1~{a}~{R},7~{b}~{S})-5-fluoranyl-2,2-bis(oxidanyl)-1~{a},7~{b}-dihydro-1~{H}-cyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
7TF7
 
 | | S. aureus GS(12) - apo | | Descriptor: | Glutamine synthetase | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
7AHM
 
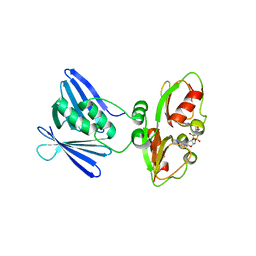 | | Low-resolution structure of the K+/H+ antiporter subunit KhtT in complex with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, K(+)/H(+) antiporter subunit KhtT | | Authors: | Cereija, T.B, Guerra, J.P, Morais-Cabral, J.H. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | c-di-AMP, a likely master regulator of bacterial K + homeostasis machinery, activates a K + exporter.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ALF
 
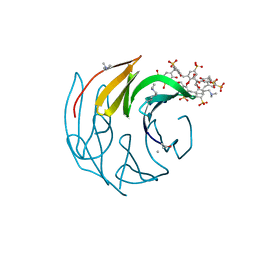 | | The dimethylated RSL - sulfonato-calix[8]arene complex, P3 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
8SSK
 
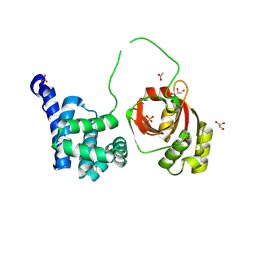 | | Citrobacter rodentium contact dependent growth inhibition (CDI) entry and toxin (CdiA-CT) domains | | Descriptor: | ACETATE ION, CALCIUM ION, Contact-dependent inhibitor A, ... | | Authors: | Cuthbert, B.J, Goulding, C.W, Hayes, C.S, Nhan, D.Q. | | Deposit date: | 2023-05-08 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Citrobacter rodentium contact dependent growth inhibition (CDI) entry and toxin (CdiA-CT) domains
To Be Published
|
|
5F4Z
 
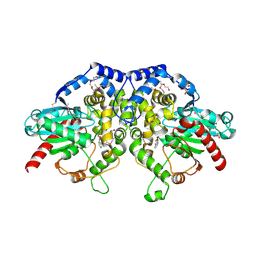 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
8Q2C
 
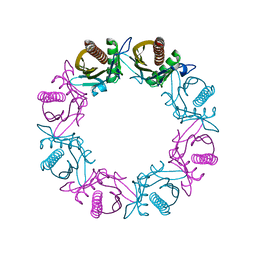 | |
5KQ2
 
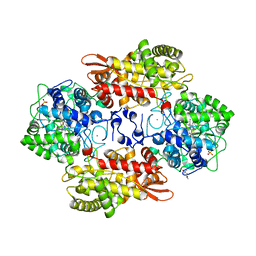 | |
7NTE
 
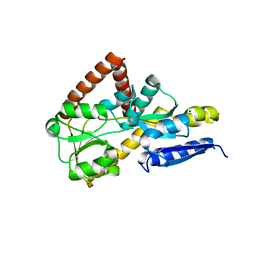 | | The structure of an open conformation of the SBP TarP_Csal | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
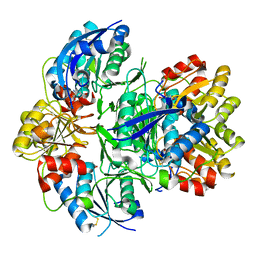
6V5F
 
 | |
8DFK
 
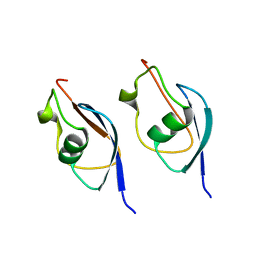 | |
5I33
 
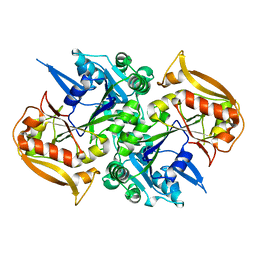 | | Unligated adenylosuccinate synthetase from Cryptococcus neoformans | | Descriptor: | Adenylosuccinate synthetase | | Authors: | Blundell, R.D, Williams, S.J, Ericsson, D, Fraser, J.A, Kobe, B. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of de Novo Adenosine Triphosphate (ATP) Biosynthesis Abolishes Virulence in Cryptococcus neoformans.
Acs Infect Dis., 2, 2016
|
|
6J1T
 
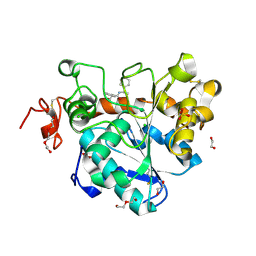 | | Crystal structure of Candida Antarctica Lipase B mutant SR with product analogue | | Descriptor: | (2S)-2-phenyl-N-[(1R)-1-phenylethyl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
7KMD
 
 | |
7AL0
 
 | | Crystal Structure of Heymonin, a Novel Frog-derived Peptide | | Descriptor: | CHLORIDE ION, Heymonin | | Authors: | Kascakova, B, Prudnikova, T, Kuta Smatanova, I, Xu, X. | | Deposit date: | 2020-10-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization and functional analysis of cathelicidin-MH, a novel frog-derived peptide with anti-septicemic properties.
Elife, 10, 2021
|
|
8PEJ
 
 | | CjGH35 with a Galactosidase Activity-Based Probe | | Descriptor: | (1~{S},2~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-06-14 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The development of a broad-spectrum retaining beta-exo-galactosidase activity-based probe.
Org.Biomol.Chem., 21, 2023
|
|
8PE3
 
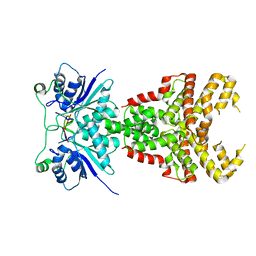 | | Structure of Csm6' from Streptococcus thermophilus in complex with cyclic hexa-adenylate (cA6) | | Descriptor: | CRISPR system endoribonuclease Csm6', Cyclic hexaadenosine monophosphate (cA6), RNA | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
7AUU
 
 | |
5IO3
 
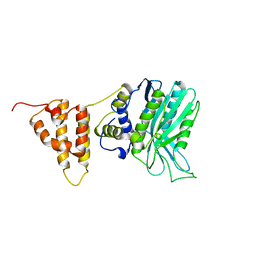 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
7AUT
 
 | |
7AUO
 
 | | Yeast Diphosphoinositol Polyphosphate Phosphohydrolase DDP1 in complex with PA-InsP8 | | Descriptor: | Diphosphoinositol polyphosphate phosphohydrolase DDP1, {[(1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl]bis[oxy(2-oxoethane-2,1-diyl)]}bis(phosphonic acid) | | Authors: | Marquez-Monino, M.A, Gonzalez, B. | | Deposit date: | 2020-11-03 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping.
Sci Adv, 7, 2021
|
|
