2IF5
 
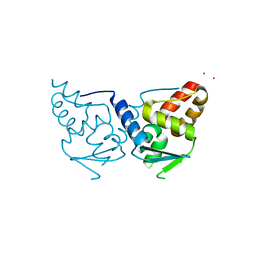 | | Structure of the POZ domain of human LRF, a master regulator of oncogenesis | | Descriptor: | PRASEODYMIUM ION, Zinc finger and BTB domain-containing protein 7A | | Authors: | Schubot, F.D, Waugh, D.S, Tropea, J. | | Deposit date: | 2006-09-20 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the POZ domain of human LRF, a master regulator of oncogenesis.
Biochem.Biophys.Res.Commun., 351, 2006
|
|
7NOW
 
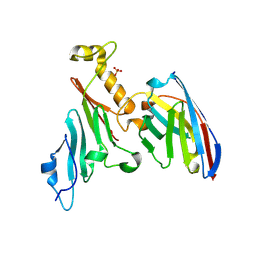 | |
1PB1
 
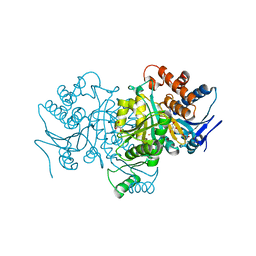 | |
1WRN
 
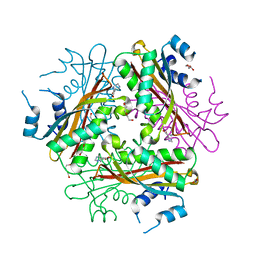 | | Metal Ion dependency of the antiterminator protein, HutP, for binding to the terminator region of hut mRNA- A structural basis | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1PB3
 
 | |
2IKQ
 
 | |
2ICT
 
 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
8UC6
 
 | | Calpain-7:IST1 Complex | | Descriptor: | Calpain-7, IST1 homolog | | Authors: | Paine, E, Whitby, F.G, Hill, C.P, Sunquist, W.I. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Elife, 12, 2023
|
|
2MDR
 
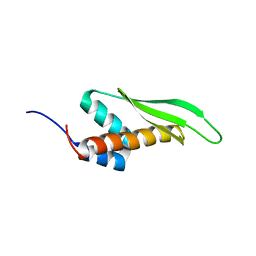 | | Solution structure of the third double-stranded RNA-binding domain (dsRBD3) of human adenosine-deaminase ADAR1 | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Barraud, P, Banerjee, S, Mohamed, W.I, Jantsch, M.F, Allain, F.H. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8U0T
 
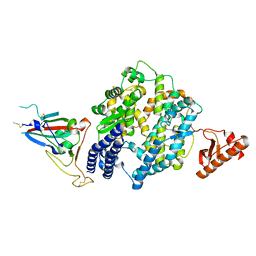 | |
8U29
 
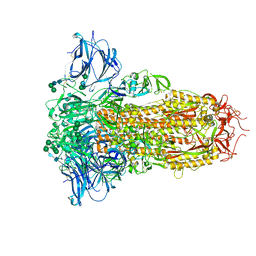 | | Prefusion structure of the PRD-0038 spike glycoprotein ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRD-0038 Spike glycoprotein, ... | | Authors: | Lee, J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-09-05 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broad receptor tropism and immunogenicity of a clade 3 sarbecovirus.
Cell Host Microbe, 31, 2023
|
|
4LYP
 
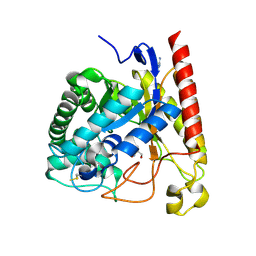 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2MVO
 
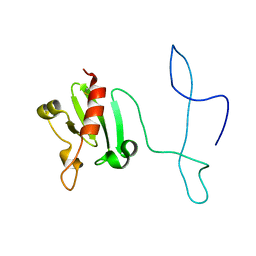 | | Solution structure of the lantibiotic self-resistance lipoprotein MlbQ from Microbispora ATCC PTA-5024 | | Descriptor: | Putative lipoprotein | | Authors: | Pozzi, R, Schwartz, P, Linke, D, Kulik, A, Nega, M, Wohlleben, W, Stegmann, E, Coles, M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Distinct mechanisms contribute to immunity in the lantibiotic NAI-107 producer strain Microbispora ATCC PTA-5024.
Environ Microbiol, 18, 2016
|
|
8U0Q
 
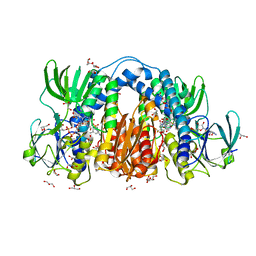 | | Co-crystal structure of optimized analog TDI-13537 provided new insights into the potency determinants of the sulfonamide inhibitor series | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dementiev, A.A, Michino, M, Vendome, J, Ginn, J, Bryk, R, Olland, A. | | Deposit date: | 2023-08-29 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Shape-Based Virtual Screening of a Billion-Compound Library Identifies Mycobacterial Lipoamide Dehydrogenase Inhibitors.
Acs Bio Med Chem Au, 3, 2023
|
|
3RUU
 
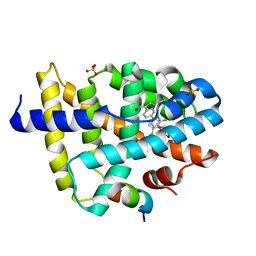 | | FXR with SRC1 and GSK237 | | Descriptor: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazol-4-yl]methoxy}phenyl)-1H-indole-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1OSJ
 
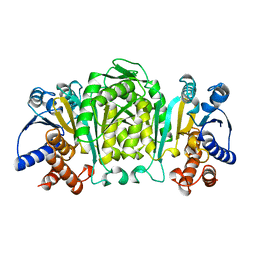 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1XGV
 
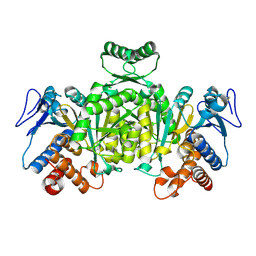 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix | | Descriptor: | Isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
1OSI
 
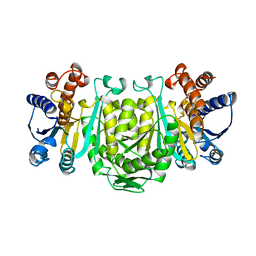 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | Deposit date: | 1996-10-22 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
2N3G
 
 | |
3RUT
 
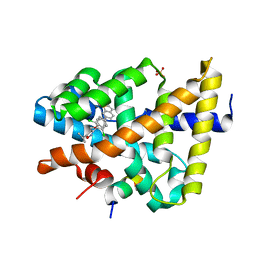 | | FXR with SRC1 and GSK359 | | Descriptor: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazol-4-yl]methoxy}phenyl)-1-benzothiophene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7N8L
 
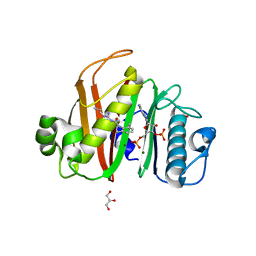 | |
7N8E
 
 | |
7N8M
 
 | |
7NQM
 
 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(2-pyrimidin-4-ylethyl)-1~{H}-indole, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
7NQO
 
 | | Mycobacterium tuberculosis Cytochrome P450 CYP121 in complex with lead compound 21 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[4-[2-(5-bromanyl-1~{H}-indol-3-yl)ethyl]pyrimidin-2-yl]morpholine, CHLORIDE ION, ... | | Authors: | Selvam, I.R. | | Deposit date: | 2021-03-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new strategy for hit generation: Novel in cellulo active inhibitors of CYP121A1 from Mycobacterium tuberculosis via a combined X-ray crystallographic and phenotypic screening approach (XP screen).
Eur.J.Med.Chem., 230, 2022
|
|
