6AP9
 
 | |
6UJV
 
 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
7CT2
 
 | |
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
9EGV
 
 | | HOIL-1 RING2 domain bound to ubiquitin | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, RanBP-type and C3HC4-type zinc finger-containing protein 1, ... | | Authors: | Wang, X.S, Lechtenberg, B.C. | | Deposit date: | 2024-11-21 | | Release date: | 2024-12-11 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The RBR E3 ubiquitin ligase HOIL-1 can ubiquitinate diverse non-protein substrates in vitro.
Life Sci Alliance, 8, 2025
|
|
2NCT
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Envelope glycoprotein gp41 | | Authors: | Jimenez, M, Nieva, J.L, Rujas, E, Partida-Hanon, A, Bruix, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface.
Sci Rep, 6, 2016
|
|
2VN8
 
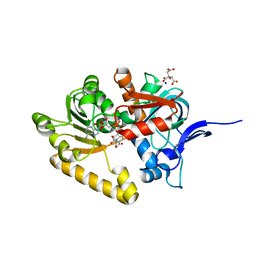 | | Crystal structure of human Reticulon 4 interacting protein 1 in complex with NADPH | | Descriptor: | CITRIC ACID, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pike, A.C.W, Guo, K, Elkins, J, Ugochukwu, E, Roos, A.K, Filippakopoulos, P, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2008-01-31 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Reticulon 4 Interacting Protein 1 in Complex with Nadph
To be Published
|
|
9EGW
 
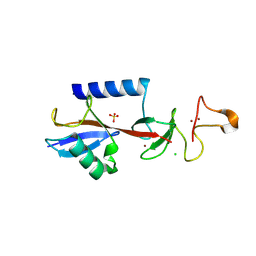 | |
5T5T
 
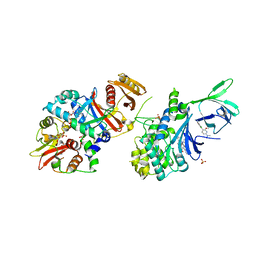 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Selective Activation of AMPK beta 1-Containing Isoforms Improves Kidney Function in a Rat Model of Diabetic Nephropathy.
J. Pharmacol. Exp. Ther., 361, 2017
|
|
5HGO
 
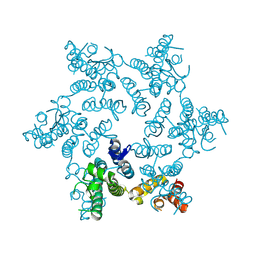 | | Hexameric HIV-1 CA R18G mutant | | Descriptor: | Capsid protein P24 | | Authors: | Jacques, D.A, James, L.C. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis.
Nature, 536, 2016
|
|
3IK9
 
 | | Human GST A1-1-GIMF with GSDHN | | Descriptor: | (S)-2-amino-5-((R)-1-(carboxymethylamino)-3-((3S,4R)-1,4-dihydroxynonan-3-ylthio)-1-oxopropan-2-ylamino)-5-oxopentanoic acid, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal.
Biochemistry, 49, 2010
|
|
4BZX
 
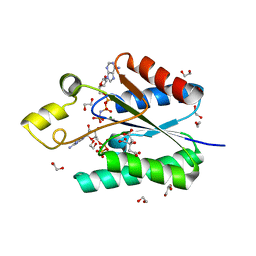 | | Structure of the Mycobacterium tuberculosis APS kinase CysC in complex with AMPPNP and APS | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-PHOSPHOSULFATE, BIFUNCTIONAL ENZYME CYSN/CYSC, ... | | Authors: | Poyraz, O, Lohkamp, B, Schnell, R, Schneider, G. | | Deposit date: | 2013-07-30 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of the Kinase Domain of the Sulfate-Activating Complex in Mycobacterium Tuberculosis.
Plos One, 10, 2015
|
|
4BVF
 
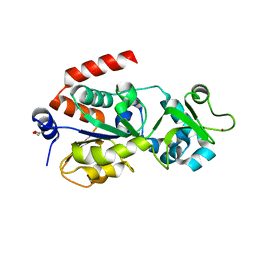 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH THIOALKYLIMIDATE FORMED FROM THIO-ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, MITOCHONDRIAL, ... | | Authors: | Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1I8X
 
 | |
6UB4
 
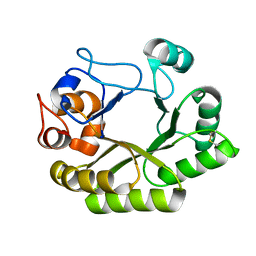 | | Crystal structure (C2 form) of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) in complex with laminaritriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCOSIDE HYDROLASE, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
7YJS
 
 | |
5DH4
 
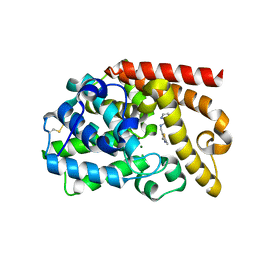 | | PDE10 complexed with 5-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 5-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6IC7
 
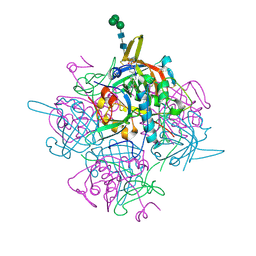 | | Human cathepsin-C in complex with dipeptidyl cyclopropyl nitrile inhibitor 3 | | Descriptor: | 1-azanyl-~{N}-[(1~{R},2~{R})-1-cyano-2-[4-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]cyclopropyl]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
5FV7
 
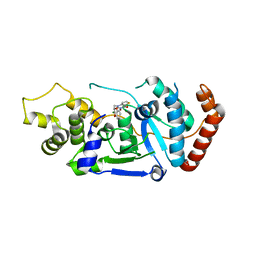 | | Human Fen1 in complex with an N-hydroxyurea compound | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, FLAP ENDONUCLEASE 1, MAGNESIUM ION | | Authors: | Exell, J.C, Thompson, M.J, Finger, L.D, Shaw, S.K, Abbott, W.M, McWhirter, C, Debreczeni, J.E, Jones, C.D, Nissink, J.W.M, Ward, T.A, Sioberg, C.W.L, Molina, D.M, Durant, S.T, Grasby, J.A. | | Deposit date: | 2016-02-03 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Cellular Active N-Hydroxyurea Fen1 Inhibitors Block Substrate Entry to the Active Site
Nat.Chem.Biol., 12, 2016
|
|
2VX0
 
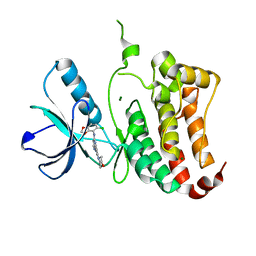 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3-MORPHOLIN-4-YLPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4OLJ
 
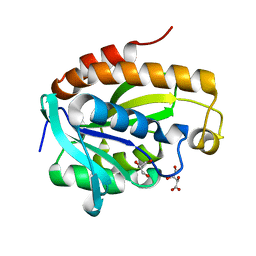 | | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA Hydrolase from Acinetobacter Baumannii at 1.49 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Sikarwar, J, Dube, D, Kaushik, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of Arg119Gln mutant of Peptidyl-tRNA hydrolase from Acinetobacter Baumannii at 1.49 A resolution
to be published
|
|
7XO6
 
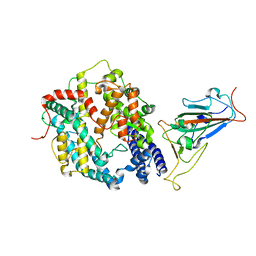 | | SARS-CoV-2 Omicron BA.1 Variant RBD with mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
4C1G
 
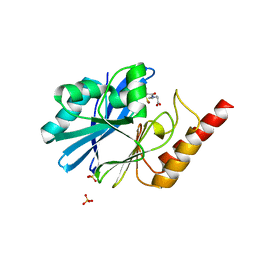 | | Crystal structure of the metallo-beta-lactamase IMP-1 with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE IMP-1, SULFATE ION, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
6B04
 
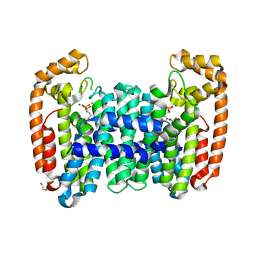 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
9IU1
 
 | | Structure of SARS-CoV-2 JN.1 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-07-20 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
