1VGJ
 
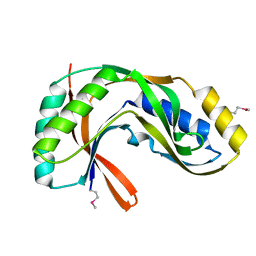 | | Crystal structure of 2'-5' RNA ligase from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH0099 | | Authors: | Yao, M, Morita, H, Okada, A, Tanaka, I. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
4FKD
 
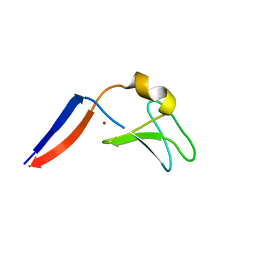 | | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta | | Descriptor: | Protein kinase C theta type, ZINC ION | | Authors: | Rahman, G.M, Shanker, S, Lewin, N.E, Prasad, B.V.V, Blumberg, P.M, Das, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta.
Biochem.J., 451, 2013
|
|
1WD1
 
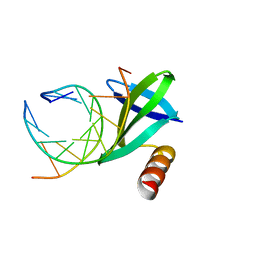 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*GP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2B5P
 
 | |
7VKE
 
 | | Crystal structure of human CD38 ECD in complex with UniDab(TM) F11A | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, CHLORIDE ION, ... | | Authors: | Schooten, W.V, Schellenberger, U, Ugamraj, H.S, Manicka, S, Bijpuria, S, Gondu, R.K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TNB-738, a biparatopic antibody, boosts intracellular NAD+ by inhibiting CD38 ecto-enzyme activity.
Mabs, 14, 2022
|
|
2IHN
 
 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
1R48
 
 | | Solution structure of the C-terminal cytoplasmic domain residues 468-497 of Escherichia coli protein ProP | | Descriptor: | Proline/betaine transporter | | Authors: | Zoetewey, D.L, Tripet, B.P, Kutateladze, T.G, Overduin, M.J, Wood, J.M, Hodges, R.S. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-terminal Antiparallel Coiled-coil Domain from Escherichia coli Osmosensor ProP.
J.Mol.Biol., 334, 2003
|
|
1UNO
 
 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3HPA
 
 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | Descriptor: | AMIDOHYDROLASE, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
3I40
 
 | | Human insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Timofeev, V.I, Bezuglov, V.V, Miroshnikov, K.A, Chuprov-Netochin, R.N, Kuranova, I.P. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1PVX
 
 | | DO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.5 | | Descriptor: | PROTEIN (ENDO-1,4-BETA-XYLANASE) | | Authors: | Rajeshkumar, P, Eswaramoorthy, S, Vithayathil, P.J, Viswamitra, M.A. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
J.Mol.Biol., 295, 2000
|
|
5YEV
 
 | | Murine DR3 death domain | | Descriptor: | SULFATE ION, TNFRSF25 death domain | | Authors: | Yin, X, Jin, T. | | Deposit date: | 2017-09-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | Descriptor: | COPPER (II) ION, Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-21 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
2JQX
 
 | | Solution structure of Malate Synthase G from joint refinement against NMR and SAXS data | | Descriptor: | Malate synthase G | | Authors: | Grishaev, A, Tugarinov, V, Kay, L.E, Trewhella, J, Bax, A. | | Deposit date: | 2007-06-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints
J.Biomol.Nmr, 40, 2008
|
|
1TBN
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
2B5Q
 
 | |
7BR9
 
 | | Crystal structure of mus musculus IRG1 | | Descriptor: | Cis-aconitate decarboxylase | | Authors: | Park, H.H, Chun, H.L. | | Deposit date: | 2020-03-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The crystal structure of mouse IRG1 suggests that cis-aconitate decarboxylase has an open and closed conformation.
Plos One, 15, 2020
|
|
5ZCI
 
 | |
6AJD
 
 | |
1TBO
 
 | | NMR STRUCTURE OF A PROTEIN KINASE C-G PHORBOL-BINDING DOMAIN, 30 STRUCTURES | | Descriptor: | PROTEIN KINASE C, GAMMA TYPE, ZINC ION | | Authors: | Xu, R.X, Pawelczyk, T, Xia, T, Brown, S.C. | | Deposit date: | 1997-04-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a protein kinase C-gamma phorbol-binding domain and study of protein-lipid micelle interactions.
Biochemistry, 36, 1997
|
|
5T0N
 
 | |
1TQN
 
 | | Crystal Structure of Human Microsomal P450 3A4 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 3A4 | | Authors: | Yano, J.K, Wester, M.R, Schoch, G.A, Griffin, K.J, Stout, C.D, Johnson, E.F. | | Deposit date: | 2004-06-17 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structure of Human Microsomal Cytochrome P450 3A4 Determined by X-ray Crystallography to 2.05-A Resolution
J.Biol.Chem., 279, 2004
|
|
2NC7
 
 | |
2NM3
 
 | | Crystal structure of dihydroneopterin aldolase from S. aureus in complex with (1S,2S)-monapterin at 1.68 angstrom resolution | | Descriptor: | ACETATE ION, D-MONAPTERIN, Dihydroneopterin aldolase | | Authors: | Blaszczyk, J, Ji, X, Yan, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the aldolase and epimerase activities of Staphylococcus aureus dihydroneopterin aldolase.
J.Mol.Biol., 368, 2007
|
|
