5WHY
 
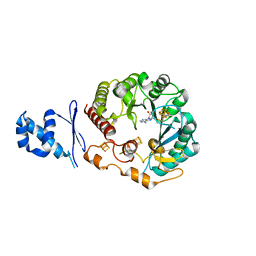 | | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, Radical SAM domain protein, ... | | Authors: | Grove, T.L, Himes, P, Bowers, A, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2017-07-18 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Structural Insights into Thioether Bond Formation in the Biosynthesis of Sactipeptides.
J. Am. Chem. Soc., 139, 2017
|
|
5V1Q
 
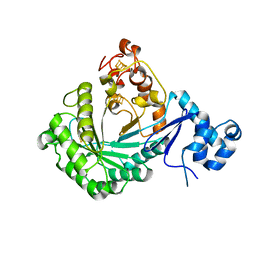 | | Crystal structure of Streptococcus suis SuiB | | Descriptor: | IRON/SULFUR CLUSTER, Radical SAM | | Authors: | Davis, K.M, Bacik, J.P, Ando, N. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VSM
 
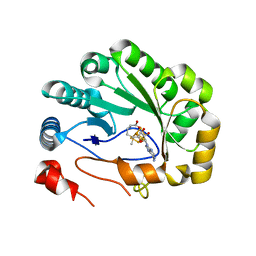 | | Crystal structure of viperin with bound [4Fe-4S] cluster, 5'-deoxyadenosine, and L-methionine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Fenwick, M.K, Li, Y, Cresswell, P, Modis, Y, Ealick, S.E. | | Deposit date: | 2017-05-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of viperin, an antiviral radical SAM enzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TH5
 
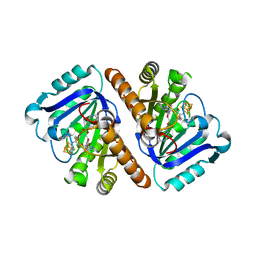 | | Crystal Structure of QueE from Bacillus subtilis with 6-carboxypterin-5'-deoxyadenosyl ester bound | | Descriptor: | 5'-O-(2-amino-4-oxo-1,4-dihydropteridine-6-carbonyl)adenosine, 7-carboxy-7-deazaguanine synthase, IRON/SULFUR CLUSTER, ... | | Authors: | Grell, T.A.J, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
J. Am. Chem. Soc., 139, 2017
|
|
4K39
 
 | | Native anSMEcpe with bound AdoMet and Cp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, Cp18Cys peptide, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K38
 
 | | Native anSMEcpe with bound AdoMet and Kp18Cys peptide | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K36
 
 | | His6 tagged anSMEcpe with bound AdoMet | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4M7S
 
 | |
7N7I
 
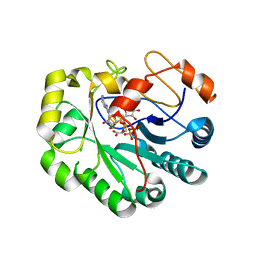 | | X-ray crystal structure of Viperin-like enzyme from Trichoderma virens | | Descriptor: | IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
7N7H
 
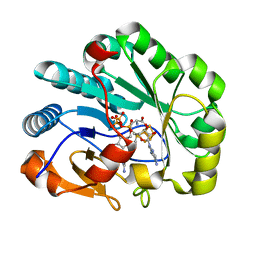 | | X-ray crystal structure of Viperin-like enzyme from Nematostella vectensis | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
4K37
 
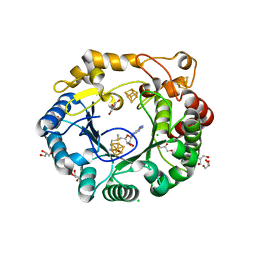 | | Native anSMEcpe with bound AdoMet | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4M7T
 
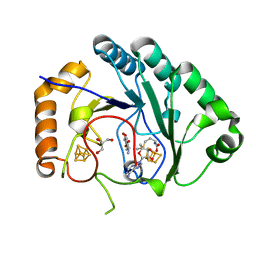 | | Crystal structure of BtrN in complex with AdoMet and 2-DOIA | | Descriptor: | (1R,2S,3S,4R,5S)-5-aminocyclohexane-1,2,3,4-tetrol, BtrN, GLYCEROL, ... | | Authors: | Drennan, C.L, Goldman, P.J. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6WTE
 
 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with cobalamin and [4Fe-4S] cluster bound | | Descriptor: | 1,2-ETHANEDIOL, B12-binding domain-containing protein, COBALAMIN, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
6WTF
 
 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with tryptophan substrate and SAM analog (aza-SAM) bound | | Descriptor: | COBALAMIN, IRON/SULFUR CLUSTER, S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
6QK7
 
 | | Elongator catalytic subcomplex Elp123 lobe | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Dauden, M.I, Jaciuk, M, Glatt, S. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of tRNA recognition by the Elongator complex.
Sci Adv, 5, 2019
|
|
6IAD
 
 | |
6IAZ
 
 | |
6IA8
 
 | |
8PU0
 
 | | Cryo-EM structure of human Elp123 in complex with tRNA, desulpho-CoA, 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, DESULFO-COENZYME A, Elongator complex protein 1, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
8PTX
 
 | | Cryo-EM structure of human Elp123 in complex with tRNA, acetyl-CoA, 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, ACETYL COENZYME *A, Elongator complex protein 1, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
8PTY
 
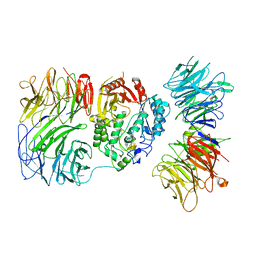 | | Cryo-EM structure of human Elp123 in complex with 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
8PTZ
 
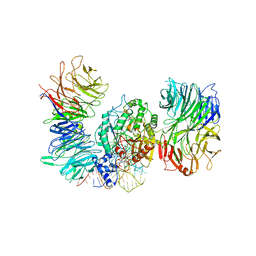 | | Cryo-EM structure of human Elp123 in complex with tRNA, S-ethyl-CoA, 5'-deoxyadenosine and methionine | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Abbassi, N, Jaciuk, M, Lin, T.-Y, Glatt, S. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of the human Elongator complex at work.
Nat Commun, 15, 2024
|
|
8ASV
 
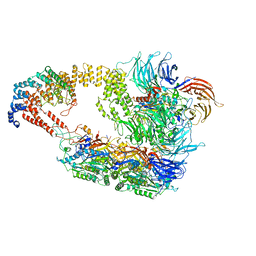 | | Cryo-EM structure of yeast Elongator complex | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASW
 
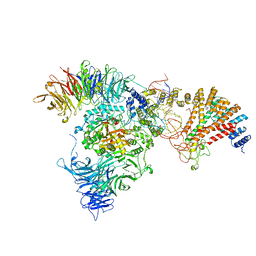 | | Cryo-EM structure of yeast Elp123 in complex with alanine tRNA | | Descriptor: | 5'-DEOXYADENOSINE, Alanine tRNA, Elongator complex protein 1, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8AVG
 
 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
