2BPG
 
 | | STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*GP*GP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*G)-3'), ... | | Authors: | Pelletier, H, Sawaya, M.R, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-05-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP.
Science, 264, 1994
|
|
8C3Q
 
 | |
8C3R
 
 | |
8C2Z
 
 | | Crystal structure of DYRK1B in complex with AZ191 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1B, MANGANESE (II) ION, N-[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]-4-(1-methylpyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural perspective on the design of selective DYRK1B inhibitors
To Be Published
|
|
3E5Y
 
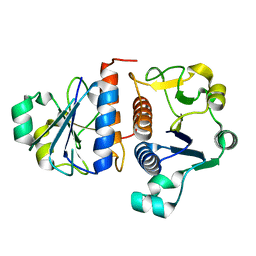 | |
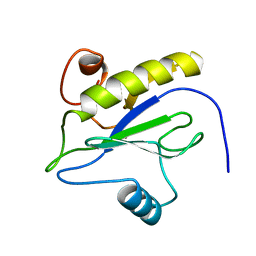
1LBA
 
 | |
6HAZ
 
 | | Crystal structure of the bromodomain of human SMARCA2 in complex with SMARCA-BD ligand | | Descriptor: | 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Bader, G, Steurer, S, Weiss-Puxbaum, A, Zoephel, A, Roy, M, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6ERJ
 
 | | Self-complemented FimA subunit from Salmonella enterica | | Descriptor: | ACETIC ACID, Type-1 fimbrial protein, a chain | | Authors: | Zyla, D.S, Prota, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 294, 2019
|
|
3FS0
 
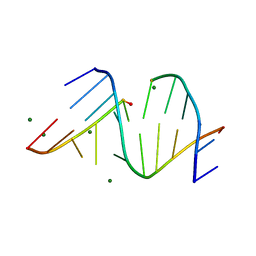 | |
3FTM
 
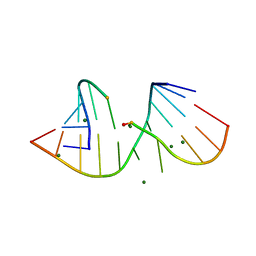 | |
2YO2
 
 | |
2YO1
 
 | |
2YO0
 
 | |
2YNZ
 
 | |
2YO3
 
 | |
1JTW
 
 | |
6IVS
 
 | |
8C3G
 
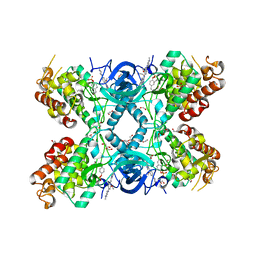 | | Crystal structure of DYRK1A in complex with AZ191 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, N-[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]-4-(1-methylpyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspective on the design of selective DYRK1B inhibitors
To Be Published
|
|
6CD0
 
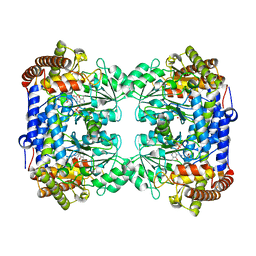 | | Crystal structure of Medicago truncatula serine hydroxymethyltransferase 3 (MtSHMT3), PLP-internal aldimine and apo form | | Descriptor: | ACETATE ION, FORMIC ACID, Serine hydroxymethyltransferase | | Authors: | Ruszkowski, M, Sekula, B, Ruszkowska, A, Dauter, Z. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Chloroplastic Serine Hydroxymethyltransferase FromMedicago truncatula: A Structural Characterization.
Front Plant Sci, 9, 2018
|
|
5E0J
 
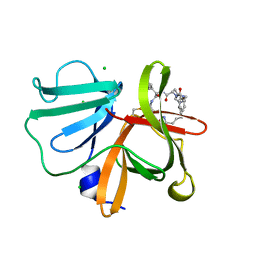 | | 1.20 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (21-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(12~{S},15~{S},18~{S})-15-(cyclohexylmethyl)-12-(hydroxymethyl)-9,14,17-tris(oxidanylidene)-1,8,13,16,21,22-hexazabicyclo[18.2.1]tricosa-20(23),21-dien-18-yl]carbamate, CHLORIDE ION, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
8SDA
 
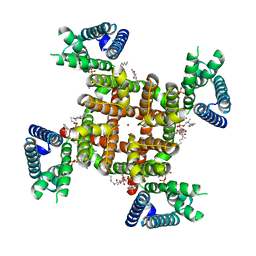 | | CryoEM structure of rat Kv2.1(1-598) L403A mutant in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
8SD3
 
 | | CryoEM structure of rat Kv2.1(1-598) wild type in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel subfamily B member 1 | | Authors: | Tan, X, Swartz, K.J. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Inactivation of the Kv2.1 channel through electromechanical coupling.
Nature, 622, 2023
|
|
5E0G
 
 | | 1.20 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (17-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(8~{S},11~{S},14~{S})-8-(hydroxymethyl)-11-(2-methylpropyl)-5,10,13-tris(oxidanylidene)-1,4,9,12,17,18-hexazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate, CHLORIDE ION, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
5E0H
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic (18-mer) inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, GLYCEROL, Norovirus 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Weerawarna, P.M, Kim, Y, Kankanamalage, A.C.G, Damalanka, V.C, Lushington, G.H, Alliston, K.R, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-09-28 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and synthesis of triazole-based macrocyclic inhibitors of norovirus protease: Structural, biochemical, spectroscopic, and antiviral studies.
Eur.J.Med.Chem., 119, 2016
|
|
1NGM
 
 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
