1KXT
 
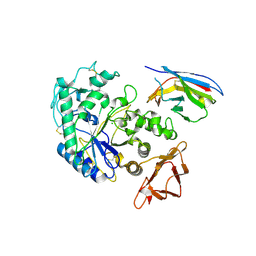 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CALCIUM ION, ... | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
2N1Q
 
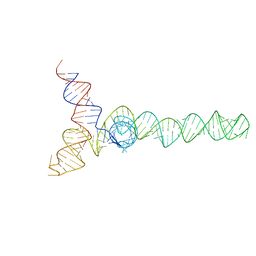 | | HIV-1 Core Packaging Signal | | Descriptor: | RNA_(155-MER) | | Authors: | Keane, S.C, Summers, M.F. | | Deposit date: | 2015-04-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RNA structure. Structure of the HIV-1 RNA packaging signal.
Science, 348, 2015
|
|
1XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, 5 STRUCTURES | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-16 | | Release date: | 1999-07-21 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
2N62
 
 | | ddFLN5+110 | | Descriptor: | gelation factor, secretion monitor chimera | | Authors: | Cabrita, L.D, Cassaignau, A.M.E, Launay, H.M.M, Waudby, C.A, Camilloni, C, Robertson, A.L, Wang, X, Wlodarski, T, Wentink, A.S, Vendruscolo, M, Dobson, C.M, Christodoulou, J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1HN1
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION, SODIUM ION | | Authors: | Juers, D.H, Matthews, B.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible lattice repacking illustrates the temperature dependence of macromolecular interactions.
J.Mol.Biol., 311, 2001
|
|
2N8Q
 
 | |
2NAT
 
 | |
2NAU
 
 | |
2OAU
 
 | | Mechanosensitive Channel of Small Conductance (MscS) | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Rees, D.C, Bass, R.B, Steinbacher, S, Strop, P, Barclay, M.T. | | Deposit date: | 2006-12-17 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS
CURRENT TOPICS IN MEMBRANES, 58, 2007
|
|
1PMT
 
 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-03-23 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
1QME
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1QMF
 
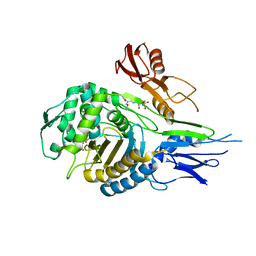 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1QJ3
 
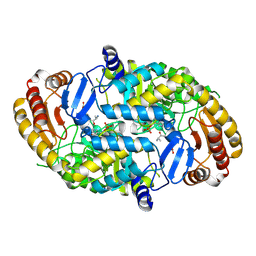 | | Crystal structure of 7,8-diaminopelargonic acid synthase in complex with 7-keto-8-aminopelargonic acid | | Descriptor: | 7,8-DIAMINOPELARGONIC ACID SYNTHASE, 7-KETO-8-AMINOPELARGONIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1999-06-21 | | Release date: | 2000-06-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Diaminopelargonic Acid Synthase; Evolutionary Relationships between Pyridoxal-5'-Phosphate Dependent Enzymes
J.Mol.Biol., 291, 1999
|
|
1R4W
 
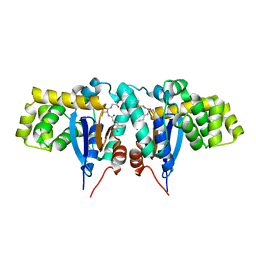 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1S9E
 
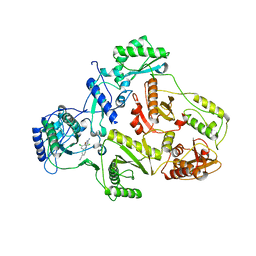 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
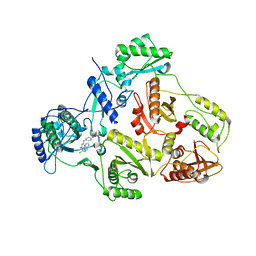 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S6Q
 
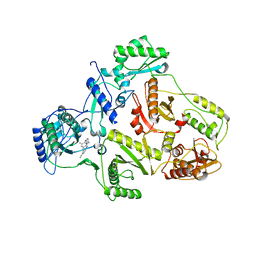 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1PMD
 
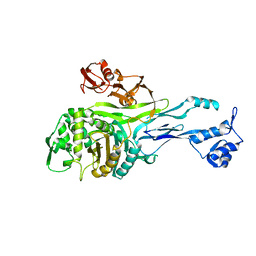 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PEPTIDOGLYCAN SYNTHESIS MULTIFUNCTIONAL ENZYME | | Authors: | Pares, S, Mouz, N, Dideberg, O. | | Deposit date: | 1996-02-05 | | Release date: | 1997-02-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme.
Nat.Struct.Biol., 3, 1996
|
|
1SUQ
 
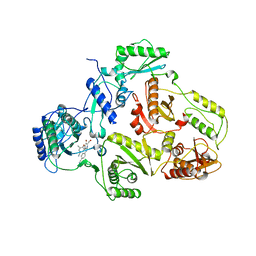 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | Descriptor: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1SV5
 
 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-27 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1SQF
 
 | | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | | Descriptor: | S-ADENOSYLMETHIONINE, SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
1E7Z
 
 | | Crystal structure of the EMAP2/RNA binding domain of the p43 protein from human aminoacyl-tRNA synthetase complex | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II, MERCURY (II) ION | | Authors: | Pasqualato, S, Kerjan, P, Renault, L, Menetrey, J, Mirande, M, Cherfils, J. | | Deposit date: | 2000-09-13 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the Emapii Domain of Human Aminoacyl-tRNA Synthetase Complex Reveals Evolutionary Dimeric Mimicry
Embo J., 20, 2001
|
|
1FL0
 
 | | CRYSTAL STRUCTURE OF THE EMAP2/RNA-BINDING DOMAIN OF THE P43 PROTEIN FROM HUMAN AMINOACYL-TRNA SYNTHETASE COMPLEX | | Descriptor: | ENDOTHELIAL-MONOCYTE ACTIVATING POLYPEPTIDE II | | Authors: | Renault, L, Kerjan, P, Pasqualato, S, Menetrey, J, Robinson, J.-C, Kawaguchi, S, Vassylyev, D.G, Yokoyama, S, Mirande, M, Cherfils, J. | | Deposit date: | 2000-08-11 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry.
EMBO J., 20, 2001
|
|
1FN4
 
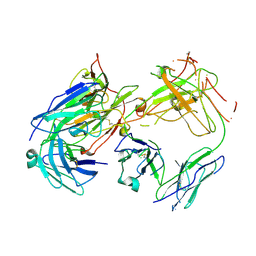 | | CRYSTAL STRUCTURE OF FAB198, AN EFFICIENT PROTECTOR OF ACETYLCHOLINE RECEPTOR AGAINST MYASTHENOGENIC ANTIBODIES | | Descriptor: | MONOCLONAL ANTIBODY AGAINST ACETYLCHOLINE RECEPTOR | | Authors: | Poulas, K, Eliopoulos, E, Vatzaki, E, Navaza, J, Kontou, M. | | Deposit date: | 2000-08-21 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Fab198, an efficient protector of the acetylcholine receptor against myasthenogenic antibodies.
Eur.J.Biochem., 268, 2001
|
|
1G6I
 
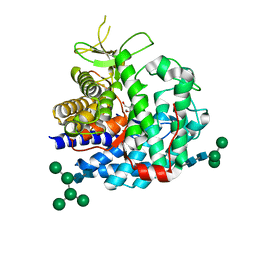 | | Crystal structure of the yeast alpha-1,2-mannosidase with bound 1-deoxymannojirimycin at 1.59 A resolution | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Herscovics, A, Lipari, F, Sleno, B, Romera, P.A, Vallee, F, Yip, P, Howell, P.A. | | Deposit date: | 2000-11-06 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and function of Class I a1,2-mannosidases involved
in glycoprotein biosynthesis.
CARBOHYDRATE BIOENGINEERING. INTERDISCIPLINARY APPROACHES., 2002
|
|
