5SCB
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 28 | | Descriptor: | (3R)-1-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carboxylic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCG
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 101 | | Descriptor: | (3R)-1-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carboxylic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCL
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 1 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ALIZARIN RED, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCD
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 58 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SC9
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 29 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 1-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-4-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCE
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 55 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-amino-4-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCA
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 36 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-[(3R)-1-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carbonyl]-L-aspartic acid, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCH
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 100 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SC8
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 17 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-[(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)amino]pentanoic acid, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5PZM
 
 | |
5PGW
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3- HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE | | Descriptor: | 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PZN
 
 | |
5PZO
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE C316N IN COMPLEX WITH 2-(4-FLUOROPHENYL)-N-METHYL-5-[3-({2-METHYL-1-OXO-1-[(1,3,4-THIADIAZOL-2-YL)AMINO]PROPAN-2-YL}CARBAMOYL)PHENYL]-1-BENZOFURAN-3-CARBOXAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-(4-fluorophenyl)-N-methyl-5-[3-({2-methyl-1-oxo-1-[(1,3,4-thiadiazol-2-yl)amino]propan-2-yl}carbamoyl)phenyl]-1-benzofuran-3-carboxamide, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
5PZK
 
 | |
5PZL
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 2-({3-[1-(2-CYCLOPROPYLETHYL)-6-FLUORO-4-HYDROXY-2-OXO-1,2-DIHYDROQUINOLIN-3-YL]-1,1-DIOXO-1,4-DIHYDRO-1LAMBDA~6~,2,4-BENZOTHIADIAZIN-7-YL}OXY)ACETAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-({3-[1-(2-cyclopropylethyl)-6-fluoro-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxo-1,4-dihydro-1lambda~6~,2,4-benzothiadiazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
5PZP
 
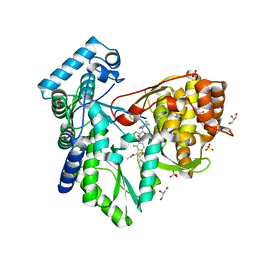 | |
5SW8
 
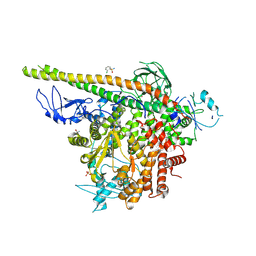 | | Crystal structure of PI3Kalpha in complex with fragments 7 and 11 | | Descriptor: | 2-CHLOROBENZENESULFONAMIDE, 2H-indazol-5-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWP
 
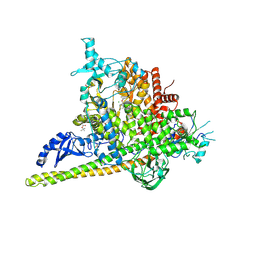 | | Crystal Structure of PI3Kalpha in complex with fragments 6 and 24 | | Descriptor: | 2-methylcyclohexane-1,3-dione, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SX5
 
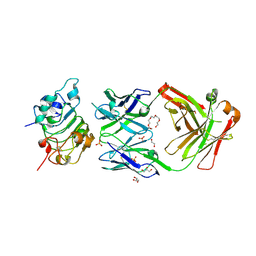 | |
5T15
 
 | | Structural basis for gating and activation of RyR1 (30 uM Ca2+ dataset, all particles) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T0L
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF and 5-(4-(3,4-dichlorophenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-15) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-[4-(3,4-dichlorophenyl)piperazin-1-yl]pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Chen, Z, Lu, H, Li, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Potent and Selective Leads against Toxoplasma gondii Dihydrofolate Reductase via Structure-Based Design.
ACS Med Chem Lett, 7, 2016
|
|
5SXB
 
 | | Crystal Structure of PI3Kalpha in complex with fragment 23 | | Descriptor: | ISATOIC ANHYDRIDE, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SXD
 
 | | Crystal Structure of PI3Kalpha in complex with fragment 22 | | Descriptor: | 2-methoxybenzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
5SWR
 
 | | Crystal Structure of PI3Kalpha in complex with fragments 20 and 26 | | Descriptor: | 2-HYDROXYBENZOIC ACID, 6-hydroxy-3,4-dihydronaphthalen-1(2H)-one, CHLORIDE ION, ... | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
