7P2J
 
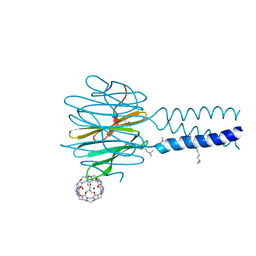 | | Dimethylated fusion protein of RSL and trimeric coiled coil (4dzn) in complex with cucurbit[7]uril, H3 sheet assembly | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, beta-D-mannopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
2OAW
 
 | | Structure of SHH variant of "Bergerac" chimera of spectrin SH3 | | Descriptor: | CHLORIDE ION, Spectrin alpha chain, brain | | Authors: | Gabdoulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Viguera, A.R, Serrano, L, Filimonov, V.V. | | Deposit date: | 2006-12-18 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic studies of Bergerac-SH3 chimeras.
Biophys.Chem., 139, 2009
|
|
1RXJ
 
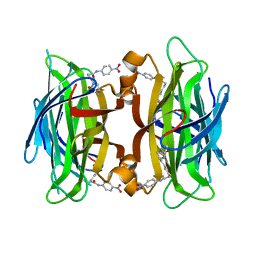 | | Crystal structure of streptavidin mutant (M2) where the L3,4 loop was replace by that of avidin | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1HAV
 
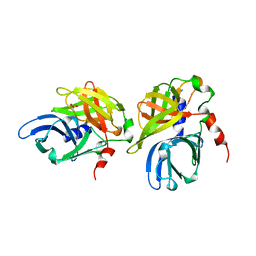 | | HEPATITIS A VIRUS 3C PROTEINASE | | Descriptor: | CHLORIDE ION, HEPATITIS A VIRUS 3C PROTEINASE | | Authors: | Bergmann, E.M, James, M.N.G. | | Deposit date: | 1996-10-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition.
J.Virol., 71, 1997
|
|
4EC2
 
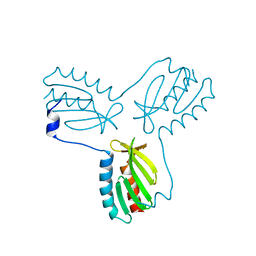 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
6Z5Q
 
 | | The RSL - sulfonato-calix[8]arene complex, P3 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5X
 
 | | The RSL - sulfonato-calix[8]arene complex, P213 form, acetate pH 4.8 | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, SULFATE ION, ... | | Authors: | Engilberge, S, Ramberg, K, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5G
 
 | | The RSL - sulfonato-calix[8]arene complex, I23 form, citrate pH 4.0, solved by S-SAD | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Engilberge, S, Ramberg, K, Crowley, P.B. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5P
 
 | | The RSL-R8 - sulfonato-calix[8]arene complex, P3 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SULFATE ION, ... | | Authors: | Ramberg, K, Skorek, T, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5M
 
 | | The RSL - sulfonato-calix[8]arene complex, I23 form, Gly-HCl pH 2.2 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5W
 
 | | The RSL - sulfonato-calix[8]arene complex, P213 form, MES pH 6.8 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Engilberge, S, Ramberg, K, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.187 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z5Z
 
 | | The RSL-R6 - sulfonato-calix[8]arene complex, P213 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, beta-D-fructopyranose, sulfonato-calix[8]arene | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z60
 
 | | The RSL - sulfonato-calix[8]arene complex, P213 form, CAPS pH 9.5 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.166 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6Z62
 
 | | The RSL - sulfonato-calix[8]arene complex, P213 form, TRIS-HCl pH 8.5 | | Descriptor: | Fucose-binding lectin protein, SULFATE ION, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-05-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.156 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
2Y26
 
 | | Transmission defective mutant of Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Sauter, C, Lorber, B, Bron, P, Trapani, S, Bergdoll, M, Marmonier, A, Schmitt-Keichinger, C, Lemaire, O, Demangeat, G, Ritzenthaler, C. | | Deposit date: | 2010-12-13 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into Viral Determinants of Nematode Mediated Grapevine Fanleaf Virus Transmission.
Plos Pathog., 7, 2011
|
|
3D8B
 
 | | Crystal structure of human fidgetin-like protein 1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Karlberg, T, Wisniewska, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human fidgetin-like protein 1.
To be Published
|
|
8H4Q
 
 | |
7UUG
 
 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with ML1006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7UUP
 
 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8PAV
 
 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | Descriptor: | 1-[3,5-bis(fluoranyl)-4-[[3-(1,3-thiazol-5-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, ... | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAW
 
 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | Descriptor: | 1-[3,5-bis(fluoranyl)-4-[[3-(1-propan-2-ylpyrazol-3-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ASPARTIC ACID, ... | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
3K5H
 
 | | Crystal structure of carboxyaminoimidazole ribonucleotide synthase from asperigillus clavatus complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosyl-aminoimidazole carboxylase | | Authors: | Thoden, J.B, Holden, H.M, Paritala, H, Firestine, S.M. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional studies of Aspergillus clavatus N(5)-carboxyaminoimidazole ribonucleotide synthetase
Biochemistry, 49, 2010
|
|
7NKW
 
 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
3KCZ
 
 | | Human poly(ADP-ribose) polymerase 2, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Karlberg, T, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Kotzsch, A, Kraulis, P, Nielsen, T.K, Moche, M, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888.
Biochemistry, 49, 2010
|
|
6YQN
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]t rideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]benzamide | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
