1Q3W
 
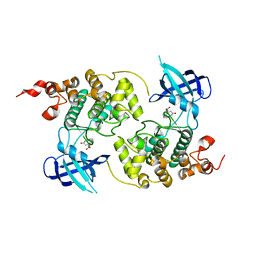 | | GSK-3 Beta complexed with Alsterpaullone | | Descriptor: | 9-NITRO-5,12-DIHYDRO-7H-BENZO[2,3]AZEPINO[4,5-B]INDOL-6-ONE, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
3BAD
 
 | |
1PUE
 
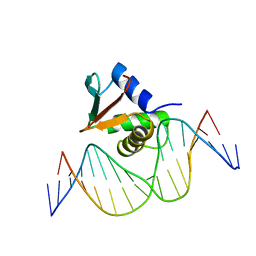 | | PU.1 ETS DOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*GP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*CP*CP*TP*TP*TP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR PU.1 (TF PU.1)) | | Authors: | Kodandapani, R, Pio, F, Ni, C.Z, Piccialli, G, Klemsz, M, McKercher, S, Maki, R.A, Ely, K.R. | | Deposit date: | 1996-07-08 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex.
Nature, 380, 1996
|
|
3BES
 
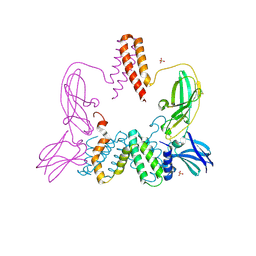 | | Structure of a Poxvirus ifngbp/ifng Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon gamma, Interferon-gamma binding protein C4R, ... | | Authors: | Nuara, A.A, Walter, M.R. | | Deposit date: | 2007-11-20 | | Release date: | 2008-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of IFN-gamma antagonism by an orthopoxvirus IFN-gamma-binding protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BAO
 
 | |
1Q41
 
 | | GSK-3 Beta complexed with Indirubin-3'-monoxime | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1POG
 
 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
1PYC
 
 | | CYP1 (HAP1) DNA-BINDING DOMAIN (RESIDUES 60-100), NMR, 15 STRUCTURES | | Descriptor: | CYP1, ZINC ION | | Authors: | Timmerman, J, Vuidepot, A.-L, Bontems, F, Lallemand, J.-Y, Gervais, M, Shechter, E, Guiard, B. | | Deposit date: | 1996-02-17 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N resonance assignment and three-dimensional structure of CYP1 (HAP1) DNA-binding domain.
J.Mol.Biol., 259, 1996
|
|
2ZQV
 
 | |
1KE8
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE | | Descriptor: | 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2ZHJ
 
 | |
2ZVU
 
 | | Crystal structure of rat heme oxygenase-1 in complex with ferrous verdoheme | | Descriptor: | 5-OXA-PROTOPORPHYRIN IX CONTAINING FE, FORMIC ACID, Heme oxygenase 1 | | Authors: | Sato, H, Sugishima, M, Fukuyama, K, Noguchi, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat haem oxygenase-1 in complex with ferrous verdohaem: presence of a hydrogen-bond network on the distal side
Biochem.J., 419, 2009
|
|
1KE6
 
 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Descriptor: | Cell division protein kinase 2, N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
2ZHI
 
 | |
2ZFN
 
 | |
1LFU
 
 | | NMR Solution Structure of the Extended PBX Homeodomain Bound to DNA | | Descriptor: | 5'-D(*GP*CP*GP*CP*AP*TP*GP*AP*TP*TP*GP*CP*CP*C)-3', 5'-D(*GP*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*CP*GP*C)-3', homeobox protein PBX1 | | Authors: | Sprules, T, Green, N, Featherstone, M, Gehring, K. | | Deposit date: | 2002-04-12 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lock and Key Binding of the HOX YPWM Peptide to the PBX Homeodomain
J.Biol.Chem., 278, 2003
|
|
1PIN
 
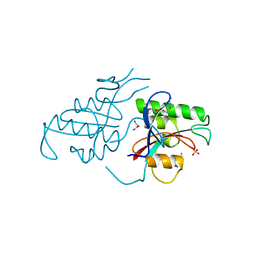 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Noel, J.P, Ranganathan, R, Hunter, T. | | Deposit date: | 1998-06-21 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
1LQC
 
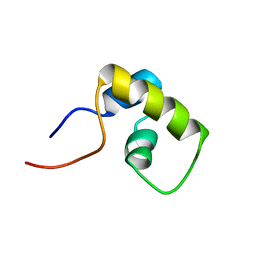 | | LAC REPRESSOR HEADPIECE (RESIDUES 1-56), NMR, 32 STRUCTURES | | Descriptor: | LAC REPRESSOR | | Authors: | Slijper, M, Bonvin, A.M.J.J, Boelens, R, Kaptein, R. | | Deposit date: | 1996-08-13 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined structure of lac repressor headpiece (1-56) determined by relaxation matrix calculations from 2D and 3D NOE data: change of tertiary structure upon binding to the lac operator.
J.Mol.Biol., 259, 1996
|
|
6WNX
 
 | | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide | | Descriptor: | Catenin beta-1, F-box/WD repeat-containing protein 11, GLYCEROL, ... | | Authors: | Ivanochko, D, Edwards, A.M, Bountra, C, Arrowsmith, C.H, Boettcher, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FBXW11-SKP1 in complex with a pSer33/pSer37 Beta-Catenin peptide
To Be Published
|
|
6WTC
 
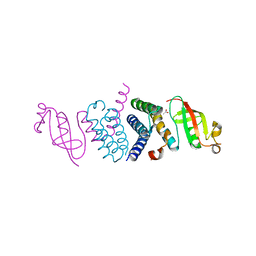 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
1KBH
 
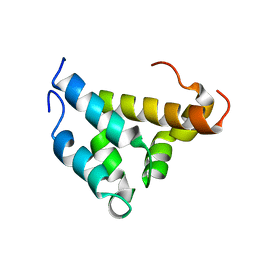 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
1K8K
 
 | | Crystal Structure of Arp2/3 Complex | | Descriptor: | ACTIN-LIKE PROTEIN 2, ACTIN-LIKE PROTEIN 3, ARP2/3 COMPLEX 16 KDA SUBUNIT, ... | | Authors: | Robinson, R.C, Turbedsky, K, Kaiser, D.A, Higgs, H.N, Marchand, J.-B, Choe, S, Pollard, T.D. | | Deposit date: | 2001-10-24 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Arp2/3 Complex
Science, 294, 2001
|
|
6WGF
 
 | | Atomic model of mutant Mcm2-7 hexamer with Mcm6 WHD truncation | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1PXK
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)pyrimidin-2-yl]-N'-hydroxyiminoformamide | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-YL]-N'-HYDROXYIMIDOFORMAMIDE | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
