1EV3
 
 | | Structure of the rhombohedral form of the M-cresol/insulin R6 hexamer | | Descriptor: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | Authors: | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | Deposit date: | 2000-04-19 | | Release date: | 2000-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | R6 hexameric insulin complexed with m-cresol or resorcinol.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EG5
 
 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-02-13 | | Release date: | 2000-04-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
3DAV
 
 | |
1EXG
 
 | | SOLUTION STRUCTURE OF A CELLULOSE BINDING DOMAIN FROM CELLULOMONAS FIMI BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | Xu, G.-Y, Ong, E, Gilkes, N.R, Kilburn, D.G, Muhandiram, D.R, Harris-Brandts, M, Carver, J.P, Kay, L.E, Harvey, T.S. | | Deposit date: | 1995-03-14 | | Release date: | 1995-06-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
3QJZ
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
1E4R
 
 | | Solution structure of the mouse defensin mBD-8 | | Descriptor: | Beta-defensin 8 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
3DB5
 
 | | Crystal structure of methyltransferase domain of human PR domain-containing protein 4 | | Descriptor: | PR domain zinc finger protein 4 | | Authors: | Amaya, M.F, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Plotnikov, A.N, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of methyltransferase domain of human PR domain-containing protein 4.
To be Published
|
|
1EGK
 
 | | CRYSTAL STRUCTURE OF A NUCLEIC ACID FOUR-WAY JUNCTION | | Descriptor: | 10-23 DNA ENZYME, MAGNESIUM ION, RNA (5'-R(*AP*GP*GP*AP*GP*AP*GP*AP*GP*AP*UP*GP*GP*GP*UP*GP*CP*GP*AP*G)-3') | | Authors: | Nowakowski, J, Shim, P.J, Stout, C.D, Joyce, G.F. | | Deposit date: | 2000-02-15 | | Release date: | 2000-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Alternative conformations of a nucleic acid four-way junction.
J.Mol.Biol., 300, 2000
|
|
3UJ2
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE | | Descriptor: | Enolase 1, MAGNESIUM ION, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE
to be published
|
|
1E9J
 
 | | SOLUTION STRUCTURE OF THE A-SUBUNIT OF HUMAN CHORIONIC GONADOTROPIN [INCLUDING A SINGLE GLCNAC RESIDUE AT ASN52 AND ASN78] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHORIONIC GONADOTROPIN | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, van Kuik, J.A, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-10-18 | | Release date: | 2002-07-25 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of the N-linked glycans on the 3D structure of the free alpha-subunit of human chorionic gonadotropin.
Biochemistry, 39, 2000
|
|
3QL9
 
 | |
1EH1
 
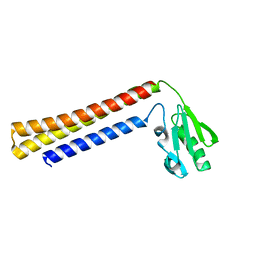 | | RIBOSOME RECYCLING FACTOR FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Toyoda, T, Tin, O.F, Ito, K, Fujiwara, T, Kumasaka, T, Yamamoto, M, Garber, M.B, Nakamura, Y. | | Deposit date: | 2000-02-18 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch.
RNA, 6, 2000
|
|
1EBA
 
 | |
1EH9
 
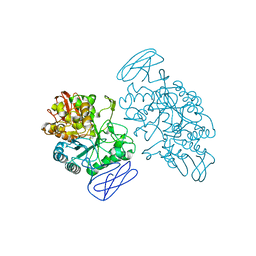 | | CRYSTAL STRUCTURE OF SULFOLOBUS SOLFATARICUS GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Descriptor: | GLYCOSYLTREHALOSE TREHALOHYDROLASE | | Authors: | Feese, M.D, Kato, Y, Tamada, T, Kato, M, Komeda, T, Kobayashi, K, Kuroki, R. | | Deposit date: | 2000-02-19 | | Release date: | 2001-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glycosyltrehalose trehalohydrolase from the hyperthermophilic archaeum Sulfolobus solfataricus.
J.Mol.Biol., 301, 2000
|
|
1EBZ
 
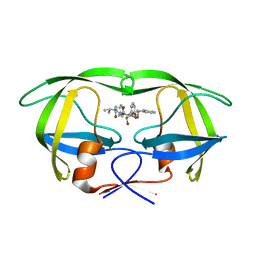 | | HIV-1 protease in complex with the inhibitor BEA388 | | Descriptor: | HIV-1 PROTEASE, [5-(2-HYDROXY-INDAN-1-YLCARBAMOYL)-3,4-DIHYDROXY-2,5-[DIBENZYL-OXY]-PENTANOYL]-VALINYL-AMIDO-METHANE | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EIE
 
 | |
1ECZ
 
 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, octyl beta-D-glucopyranoside | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
1ED7
 
 | | SOLUTION STRUCTURE OF THE CHITIN-BINDING DOMAIN OF BACILLUS CIRCULANS WL-12 CHITINASE A1 | | Descriptor: | CHITINASE A1 | | Authors: | Ikegami, T, Okada, T, Hashimoto, M, Seino, S, Watanabe, T, Shirakawa, M. | | Deposit date: | 2000-01-27 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.
J.Biol.Chem., 275, 2000
|
|
3QGJ
 
 | | 1.3A Structure of alpha-Lytic Protease Bound to Ac-AlaAlaPro-Alanal | | Descriptor: | 1,2-ETHANEDIOL, Ac-AlaAlaPro-Alanal peptide, Alpha-lytic protease, ... | | Authors: | Everill, P, Meinke, G, Bohm, A, Bachovchin, W. | | Deposit date: | 2011-01-24 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substrate Binding Defines Ser195 Position in the Catalytic Triad of Serine Proteases, Not His57 protonation
To be Published
|
|
3DDL
 
 | | Crystallographic Structure of Xanthorhodopsin, a Light-Driven Ion Pump with Dual Chromophore | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, RETINAL, ... | | Authors: | Stagno, J, Luecke, H, Schobert, B, Lanyi, J.K, Imasheva, E.S, Wang, J.M, Balashov, S.P. | | Deposit date: | 2008-06-05 | | Release date: | 2008-10-14 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1EJ4
 
 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
1EEA
 
 | | Acetylcholinesterase | | Descriptor: | PROTEIN (ACETYLCHOLINESTERASE) | | Authors: | Raves, M.L, Giles, K, Schrag, J.D, Schmid, M.F, Phillips Jr, G.N, Wah, C, Howard, A.J, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary Structure of Tetrameric Acetylcholinesterase
Structure and Function of Cholinesterases and Related Proteins, 1998
|
|
1EJE
 
 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
1EET
 
 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
