7ZSA
 
 | |
1LD8
 
 | | Co-crystal structure of Human Farnesyltransferase with farnesyldiphosphate and inhibitor compound 49 | | Descriptor: | (20S)-19,20,21,22-TETRAHYDRO-19-OXO-5H-18,20-ETHANO-12,14-ETHENO-6,10-METHENO-18H-BENZ[D]IMIDAZO[4,3-K][1,6,9,12]OXATRI AZA-CYCLOOCTADECOSINE-9-CARBONITRILE, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Taylor, J.S, Terry, K.L, Beese, L.S. | | Deposit date: | 2002-04-08 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 3-Aminopyrrolidinone farnesyltransferase inhibitors: design of macrocyclic compounds with improved pharmacokinetics and excellent cell potency.
J.Med.Chem., 45, 2002
|
|
7ZS9
 
 | |
1IHC
 
 | | X-ray Structure of Gephyrin N-terminal Domain | | Descriptor: | Gephyrin | | Authors: | Sola, M, Kneussel, M, Heck, I.S, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-04-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the trimeric N-terminal domain of gephyrin.
J.Biol.Chem., 276, 2001
|
|
1L9H
 
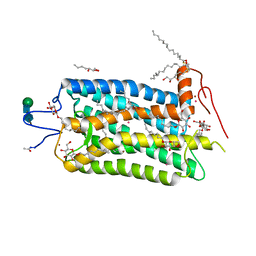 | | Crystal structure of bovine rhodopsin at 2.6 angstroms RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Fujiyoshi, Y, Silow, M, Navarro, J, Landau, E.M, Shichida, Y. | | Deposit date: | 2002-03-23 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8B0X
 
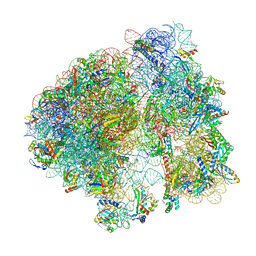 | | Translating 70S ribosome in the unrotated state (P and E, tRNAs) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fromm, S.A, O'Connor, K.M, Purdy, M, Bhatt, P.R, Loughran, G, Atkins, J.F, Jomaa, A, Mattei, S. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | The translating bacterial ribosome at 1.55 angstrom resolution generated by cryo-EM imaging services.
Nat Commun, 14, 2023
|
|
1IDH
 
 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
1LKT
 
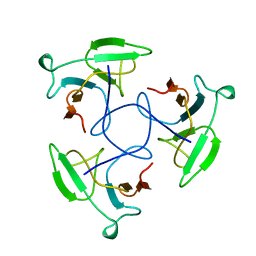 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
1LJZ
 
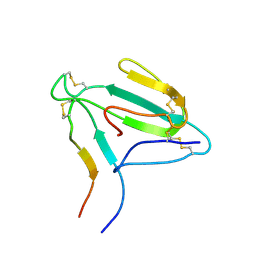 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Eisenstein, M, Chill, J.H, Anglister, J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-07-17 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1I39
 
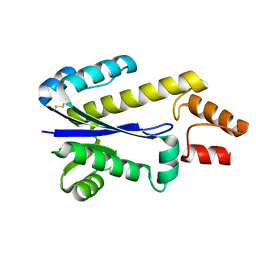 | | RNASE HII FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | RIBONUCLEASE HII | | Authors: | Chapados, B.R, Chai, Q, Hosfield, D.J, Qiu, J, Shen, B, Tainer, J.A. | | Deposit date: | 2001-02-13 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication.
J.Mol.Biol., 307, 2001
|
|
1I4D
 
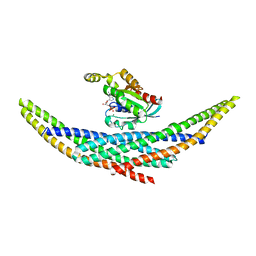 | | CRYSTAL STRUCTURE ANALYSIS OF RAC1-GDP COMPLEXED WITH ARFAPTIN (P21) | | Descriptor: | ARFAPTIN 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tarricone, C, Xiao, B, Justin, N, Gamblin, S.J, Smerdon, S.J. | | Deposit date: | 2001-02-20 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways.
Nature, 411, 2001
|
|
1LN6
 
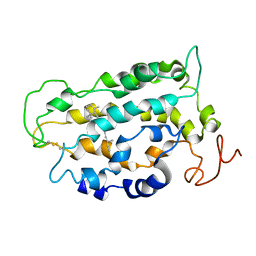 | | STRUCTURE OF BOVINE RHODOPSIN (Metarhodopsin II) | | Descriptor: | RETINAL, RHODOPSIN | | Authors: | Choi, G, Landin, J, Galan, J.F, Birge, R.R, Albert, A.D, Yeagle, P.L. | | Deposit date: | 2002-05-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin.
Biochemistry, 41, 2002
|
|
7PA2
 
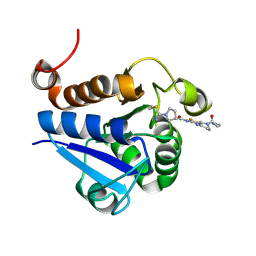 | | PARK7 with inhibitor 8RK64 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
7PA3
 
 | | PARK7 with covalent inhibitor JYQ-88 | | Descriptor: | (3~{S})-~{N}-[5-[2-[(azanylidene-$l^{4}-azanylidene)amino]ethanoyl]-6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-2-yl]-1-(iminomethyl)pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Kim, R.Q, Jia, Y, Sapmaz, A, Geurink, P.P. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput.
J.Med.Chem., 65, 2022
|
|
7Q49
 
 | | Local refinement structure of the N-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ZINC ION, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7QNG
 
 | | Structure of a MHC I-Tapasin-ERp57 complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Mueller, I.K, Thomas, C, Trowitzsch, S, Tampe, R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Nat Commun, 13, 2022
|
|
7Q3Y
 
 | | Structure of full-length, monomeric, soluble somatic angiotensin I-converting enzyme showing the N- and C-terminal ellipsoid domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
8A3V
 
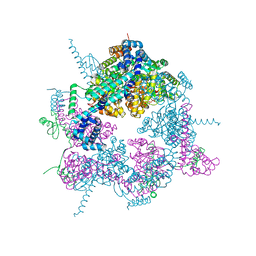 | | Crystal structure of the Vibrio cholerae replicative helicase (VcDnaB) in complex with its loader protein (VcDciA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DUF721 domain-containing protein, MAGNESIUM ION, ... | | Authors: | Walbott, H, Quevillon-Cheruel, S, Cargemel, C. | | Deposit date: | 2022-06-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The LH-DH module of bacterial replicative helicases is the common binding site for DciA and other helicase loaders.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Q4C
 
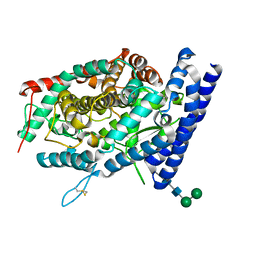 | | Local refinement structure of the C-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
1I9X
 
 | |
7Q4D
 
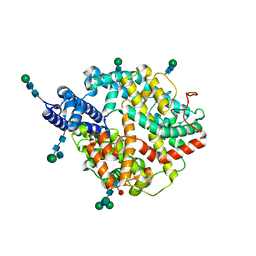 | | Local refinement structure of the two interacting N-domains of full-length, dimeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
7Q4E
 
 | | Local refinement structure of a single N-domain of full-length, dimeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
8ALZ
 
 | | Cryo-EM structure of ASCC3 in complex with ASC1 | | Descriptor: | Activating signal cointegrator 1, Activating signal cointegrator 1 complex subunit 3, ZINC ION | | Authors: | Jia, J, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of ASCC3 in complex with ASC1
Nat Commun, 2023
|
|
7QGG
 
 | | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Pulk, A, Kipper, K, Mansour, A. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state.
J.Mol.Biol., 434, 2022
|
|
1N7T
 
 | | ERBIN PDZ domain bound to a phage-derived peptide | | Descriptor: | 99-mer peptide of densin-180-like protein, phage-derived peptide | | Authors: | Skelton, N.J, Koehler, M.F.T, Zobel, K, Wong, W.L, Yeh, S, Pisabarro, M.T, Yin, J.P, Lasky, L.A, Sidhu, S.S. | | Deposit date: | 2002-11-16 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain.
J.Biol.Chem., 278, 2003
|
|
