5L1O
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
6W3A
 
 | | Structure of unphosphorylated IRE1 in complex with G-7658 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, ~{N}-[6-methyl-5-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxy-naphthalen-1-yl]but-2-ynamide | | Authors: | Ferri, E, Wang, W, Joachim, R, Mortara, K. | | Deposit date: | 2020-03-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Activation of the IRE1 RNase through remodeling of the kinase front pocket by ATP-competitive ligands.
Nat Commun, 11, 2020
|
|
8FLO
 
 | | X-ray crystal structure of substrate free CYP124A1 from Mycobacterium Marinum | | Descriptor: | Cytochrome P450 124A1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The catalytic activity and structure of the lipid metabolizing CYP124 cytochrome P450 enzyme from Mycobacterium marinum.
Arch.Biochem.Biophys., 737, 2023
|
|
4YON
 
 | | P-Rex1:Rac1 complex | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Lucato, C.M, Whisstock, J.C, Ellisdon, A.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phosphatidylinositol (3,4,5)-Trisphosphate-dependent Rac Exchanger 1Ras-related C3 Botulinum Toxin Substrate 1 (P-Rex1Rac1) Complex Reveals the Basis of Rac1 Activation in Breast Cancer Cells.
J.Biol.Chem., 290, 2015
|
|
6W3V
 
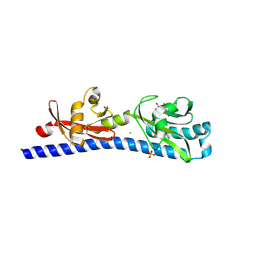 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-phenylalanine | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, PHENYLALANINE, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
6W4L
 
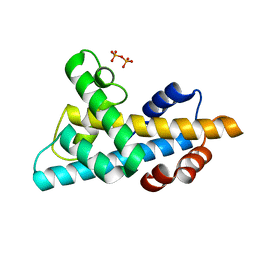 | | The crystal structure of a single chain H2B-H2A histone chimera from Xenopus laevis | | Descriptor: | Histone H2B 1.1,Histone H2A type 1, PYROPHOSPHATE | | Authors: | Warren, C, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure of a single-chain H2A/H2B dimer.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8FJO
 
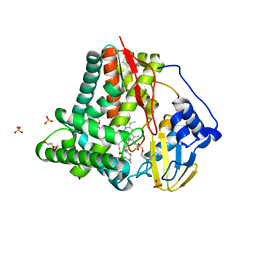 | | X-ray crystal structure of CYP124A1 from Mycobacterium Marinum in complex with farnesyl acetate | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl acetate, AMMONIUM ION, Cytochrome P450 124A1, ... | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The catalytic activity and structure of the lipid metabolizing CYP124 cytochrome P450 enzyme from Mycobacterium marinum.
Arch.Biochem.Biophys., 737, 2023
|
|
4YRL
 
 | |
5L2B
 
 | | Structure of CNTnw N149S, E332A in an outward-facing state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
6W67
 
 | |
5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L3K
 
 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a ternary complex with ADP and fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
6W7E
 
 | | K2P2.1 (TREK-1), 30 mM K+ | | Descriptor: | CADMIUM ION, DODECANE, HEXADECANE, ... | | Authors: | Lolicato, M, Minor, D.L. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | K 2P channel C-type gating involves asymmetric selectivity filter order-disorder transitions.
Sci Adv, 6, 2020
|
|
5L56
 
 | | Plexin A1 full extracellular region, domains 1 to 10, to 4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L27
 
 | | Structure of CNTnw N149L in the intermediate 1 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-07-31 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
8FKB
 
 | | X-ray crystal structure of CYP124A1 from Mycobacterium Marinum bound to Farnesol | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, Cytochrome P450 124A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The catalytic activity and structure of the lipid metabolizing CYP124 cytochrome P450 enzyme from Mycobacterium marinum.
Arch.Biochem.Biophys., 737, 2023
|
|
5L3D
 
 | | Human LSD1/CoREST: LSD1 Y761H mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
6W9H
 
 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | 4-{(3R)-3-[4-(benzyloxy)phenyl]-3-[(4-fluorophenyl)sulfonyl]pyrrolidine-1-carbonyl}-1lambda~6~-thiane-1,1-dione, Nuclear receptor ROR-gamma, Steroid receptor coactivator 1 fusion | | Authors: | Sack, J.S. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4Y5R
 
 | | Crystal Structure of a T67A MauG/pre-Methylamine Dehydrogenase Complex | | Descriptor: | CALCIUM ION, HEME C, Methylamine dehydrogenase heavy chain, ... | | Authors: | Li, C, Wilmot, C.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A T67A mutation in the proximal pocket of the high-spin heme of MauG stabilizes formation of a mixed-valent Fe(II)/Fe(III) state and enhances charge resonance stabilization of the bis-Fe(IV) state.
Biochim.Biophys.Acta, 1847, 2015
|
|
5L6I
 
 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L47
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with cyanoselenobarbital (seleniated barbiturate) | | Descriptor: | 2-[5-ethyl-2,4,6-tris(oxidanylidene)-1,3-diazinan-5-yl]ethyl selenocyanate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Reinholds Ruza, R, Fourati, Z, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
8IN9
 
 | | The structure of the GfsA KSQ-AT didomain in complex with the GfsA ACP domain | | Descriptor: | N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, Polyketide synthase | | Authors: | Chisuga, T, Murakami, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-Based Analysis of Transient Interactions between Ketosynthase-like Decarboxylase and Acyl Carrier Protein in a Loading Module of Modular Polyketide Synthase.
Acs Chem.Biol., 18, 2023
|
|
8FUO
 
 | | Fe-bound AibH1H2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amidohydrolase, FE (III) ION | | Authors: | Powell, M.M, Rittle, J. | | Deposit date: | 2023-01-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Enzymatic Hydroxylation of Aliphatic C-H Bonds by a Mn/Fe Cofactor.
J.Am.Chem.Soc., 145, 2023
|
|
6VWU
 
 | |
4YX5
 
 | | SpaO(SPOA1,2) | | Descriptor: | CHLORIDE ION, Surface presentation of antigens protein SpaO | | Authors: | Notti, R.Q, Stebbins, C.E. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9001 Å) | | Cite: | A common assembly module in injectisome and flagellar type III secretion sorting platforms.
Nat Commun, 6, 2015
|
|
