6YDW
 
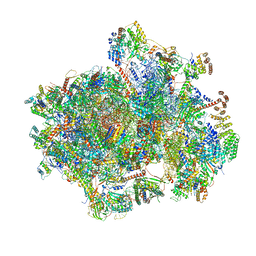 | |
6YFL
 
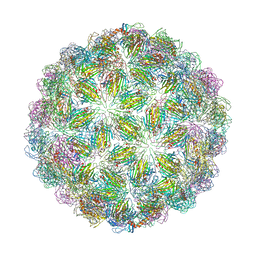 | | Virus-like particle of bacteriophage ESE020 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFT
 
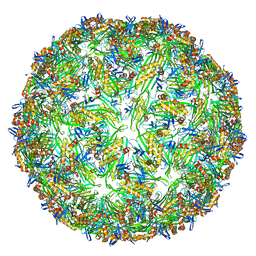 | | Virus-like particle of Wenzhou levi-like virus 1 | | Descriptor: | RNA, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
9F9S
 
 | | Yeast SDD1 Disome with Mbf1 | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S10-A, ... | | Authors: | Denk, T, Beckmann, R. | | Deposit date: | 2024-05-08 | | Release date: | 2024-12-04 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Multiprotein bridging factor 1 is required for robust activation of the integrated stress response on collided ribosomes.
Mol.Cell, 84, 2024
|
|
6YFA
 
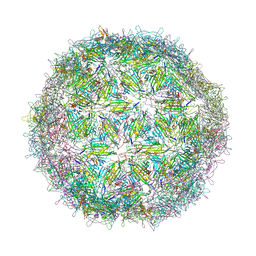 | | Virus-like particle of bacteriophage AVE015 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFH
 
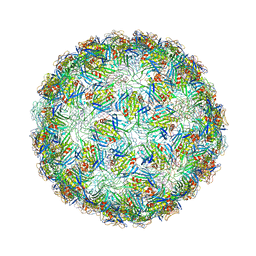 | | Virus-like particle of bacteriophage EMS014 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.893 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFQ
 
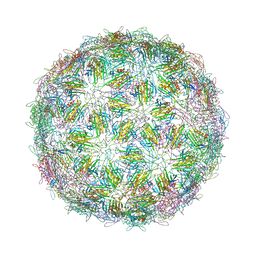 | | Virus-like particle of bacteriophage NT-214 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
4GKL
 
 | |
4GA4
 
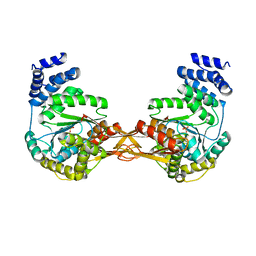 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | Descriptor: | PHOSPHATE ION, Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
9DFD
 
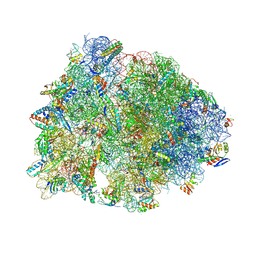 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with lasso peptide lariocidin B, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Travin, D.Y, Jangra, M, Kaur, M, Darwish, L, Koteva, K, Klepacki, D, Wang, W, Tiffany, M, Sokaribo, A, Coombes, B.K, Vazquez-Laslop, N, Wright, G.D, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome.
Res Sq, 2024
|
|
9KAY
 
 | |
4DEN
 
 | | Structural insightsinto potent, specific anti-HIV property of actinohivin; Crystal structure of actinohivin in complex with alpha(1-2) mannobiose moiety of high-mannose type glycan of gp120 | | Descriptor: | Actinohivin, POTASSIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-01-20 | | Release date: | 2012-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the specific anti-HIV property of actinohivin: structure of its complex with the alpha(1–2)mannobiose moiety of gp120
Acta Crystallogr.,Sect.D, 68, 2012
|
|
9F1B
 
 | | Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9F1C
 
 | | Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9F1D
 
 | | Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E-HYPK | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9J1M
 
 | | KU13-bond Mycobacterium tuberculosis 70S ribosome | | Descriptor: | (2~{R},3~{R},4~{R},5~{R},8~{R},10~{R},11~{R},12~{S},13~{S},14~{R})-11-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-4-[(2~{R},3~{R},4~{R},5~{S},6~{R})-6-(hydroxymethyl)-3,4-bis(oxidanyl)-5-[[4-(4-pyridin-4-yl-1,2,3-triazol-1-yl)phenyl]methoxy]oxan-2-yl]oxy-13-[(2~{R},4~{R},5~{S},6~{S})-4-methoxy-4,6-dimethyl-5-oxidanyl-oxan-2-yl]oxy-3,5,6,8,10,12,14-heptamethyl-3,10-bis(oxidanyl)-1-oxa-6-azacyclopentadecan-15-one, 16S rRNA, 23S rRNA, ... | | Authors: | Nishihara, D, Fujino, M, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2024-08-05 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Creation of a macrolide antibiotic against non-tuberculous Mycobacterium using late-stage boron-mediated aglycon delivery.
Sci Adv, 11, 2025
|
|
4GJJ
 
 | |
4G01
 
 | | ARA7-GDP-Ca2+/VPS9a | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein RABF2b, ... | | Authors: | Ihara, K. | | Deposit date: | 2012-07-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct metal recognition by guanine nucleotide-exchange factor in the initial step of the exchange reaction
Acta Crystallogr.,Sect.D, 69, 2013
|
|
9IWN
 
 | | X-ray structure of human PPARalpha ligand binding domain-intrinsic fatty acid (E. coli origin)-PGC1alpha coactivator peptide co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha, ... | | Authors: | Kamata, S, Honda, A, Yashiro, S, Komori, Y, Shimamura, A, Hosoda, A, Oyama, T, Ishii, I. | | Deposit date: | 2024-07-25 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Competitive Ligand-Induced Recruitment of Coactivators to Specific PPAR alpha / delta / gamma Ligand-Binding Domains Revealed by Dual-Emission FRET and X-Ray Diffraction of Cocrystals.
Antioxidants, 14, 2025
|
|
9IWM
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-TRAP220 coactivator peptide co-crystals obtained by cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kamata, S, Honda, A, Yashiro, S, Komori, Y, Shimamura, A, Hosoda, A, Oyama, T, Ishii, I. | | Deposit date: | 2024-07-25 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Competitive Ligand-Induced Recruitment of Coactivators to Specific PPAR alpha / delta / gamma Ligand-Binding Domains Revealed by Dual-Emission FRET and X-Ray Diffraction of Cocrystals.
Antioxidants, 14, 2025
|
|
9IWO
 
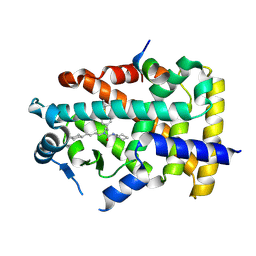 | | X-ray structure of human PPARalpha ligand binding domain-GW7647-PGC1alpha coactivator peptide co-crystals obtained by cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Kamata, S, Honda, A, Yashiro, S, Komori, Y, Shimamura, A, Hosoda, A, Oyama, T, Ishii, I. | | Deposit date: | 2024-07-25 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Competitive Ligand-Induced Recruitment of Coactivators to Specific PPAR alpha / delta / gamma Ligand-Binding Domains Revealed by Dual-Emission FRET and X-Ray Diffraction of Cocrystals.
Antioxidants, 14, 2025
|
|
4GN9
 
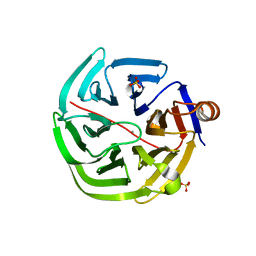 | | mouse SMP30/GNL-glucose complex | | Descriptor: | CALCIUM ION, Regucalcin, SULFATE ION, ... | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
4GPW
 
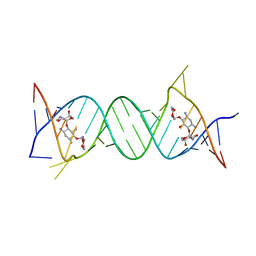 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-hydroxysisomicin (P21212 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
4GO6
 
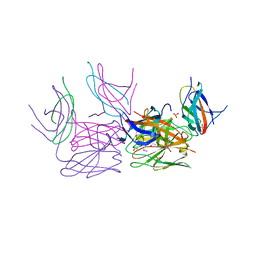 | | Crystal structure of HCF-1 self-association sequence 1 | | Descriptor: | HCF C-terminal chain 1, HCF N-terminal chain 1, SULFATE ION | | Authors: | Park, J, Lammers, F, Herr, W, Song, J. | | Deposit date: | 2012-08-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | HCF-1 self-association via an interdigitated Fn3 structure facilitates transcriptional regulatory complex formation
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GQW
 
 | |
