3VE5
 
 | |
4ZSF
 
 | | Crystal structure of pre-specific restriction endonuclease BsaWI-DNA complex | | Descriptor: | BsaWI endonuclease, CALCIUM ION, DNA, ... | | Authors: | Tamulaitiene, G, Rutkauskas, M, Grazulis, S, Siksnys, V. | | Deposit date: | 2015-05-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional significance of protein assemblies predicted by the crystal structure of the restriction endonuclease BsaWI.
Nucleic Acids Res., 43, 2015
|
|
2BZE
 
 | | NMR Structure of human RTF1 PLUS3 domain. | | Descriptor: | KIAA0252 PROTEIN | | Authors: | Truffault, V, Diercks, T, Ab, E, De Jong, R.N, Daniels, M.A, Kaptein, R, Folkers, G.E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-08-16 | | Release date: | 2007-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA Binding of the Human Rtf1 Plus3 Domain.
Structure, 16, 2008
|
|
3VDU
 
 | |
3QX3
 
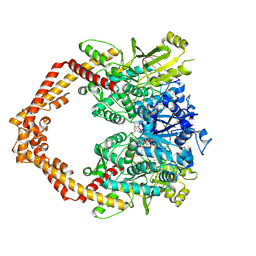 | | Human topoisomerase IIbeta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wu, C.C, Li, T.K, Farh, L, Lin, L.Y, Lin, T.S, Yu, Y.J, Yen, T.J, Chiang, C.W, Chan, N.L. | | Deposit date: | 2011-03-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide
Science, 333, 2011
|
|
5NV2
 
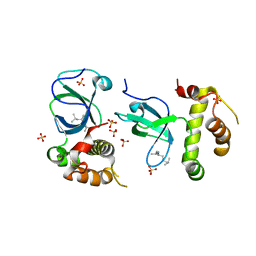 | |
5NVO
 
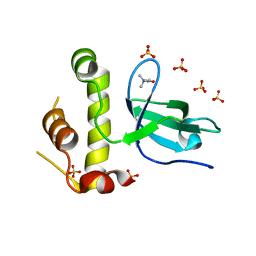 | | Human DNMT3B PWWP domain in complex with choline | | Descriptor: | CHOLINE ION, DNA (cytosine-5)-methyltransferase 3B, SULFATE ION | | Authors: | Rondelet, G, Wouters, J. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting PWWP domain of DNA methyltransferase 3B for epigenetic cancer therapy: Identification and structural characterization of new potential protein-protein interaction inhibitors
To be published
|
|
2JC5
 
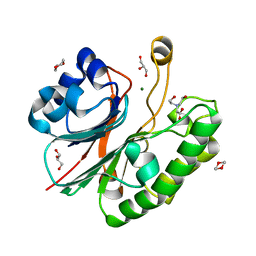 | | Apurinic Apyrimidinic (AP) endonuclease (NApe) from Neisseria Meningitidis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, BICINE, EXODEOXYRIBONUCLEASE, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G.S, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
1L9G
 
 | | CRYSTAL STRUCTURE OF URACIL-DNA GLYCOSYLASE FROM T. MARITIMA | | Descriptor: | Conserved hypothetical protein, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Rajashankar, K.R, Dodatko, T, Thirumuruhan, R.A, Sandigursky, M, Bresnik, A, Chance, M.R, Franklin, W.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-22 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of uracil-DNA glycosylase from T. Maritima
To be Published
|
|
2BM5
 
 | | The Structure of MfpA (Rv3361c, P21 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN, SULFATE ION | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2BM6
 
 | | The Structure of MfpA (Rv3361c, C2221 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | CESIUM ION, PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
4P55
 
 | |
2N7D
 
 | |
1PXE
 
 | |
2BM4
 
 | | The Structure of MfpA (Rv3361c, C2 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2ZIG
 
 | | Crystal Structure of TTHA0409, Putative DNA Modification Methylase from Thermus thermophilus HB8 | | Descriptor: | Putative modification methylase | | Authors: | Morita, R, Ishikawa, H, Nakagawa, N, Masui, R, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-15 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative DNA methylase TTHA0409 from Thermus thermophilus HB8
Proteins, 73, 2008
|
|
2BM7
 
 | | The Structure of MfpA (Rv3361c, P3221 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
5LLJ
 
 | |
3ZD9
 
 | | Potassium bound structure of E. coli ExoIX in P21 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3CJC
 
 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with DNase I and Gelsolin-segment 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3RAE
 
 | | Quinolone(Levofloxacin)-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1RV5
 
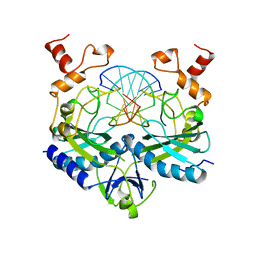 | | COMPLEX OF ECORV ENDONUCLEASE WITH D(AAAGAT)/D(ATCTT) | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*T*AP*TP*CP*TP*T)-3', ECORV ENDONUCLEASE | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 1998-06-01 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of protein-induced bending in the specificity of DNA recognition: crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + d(ATCTT).
J.Mol.Biol., 277, 1998
|
|
4KL5
 
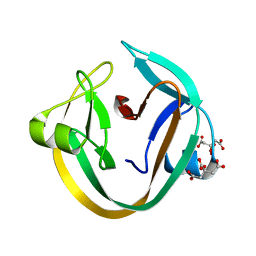 | | Crystal structure of NpuDnaE intein | | Descriptor: | CITRIC ACID, DNA polymerase III, alpha subunit, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Kajander, T, Iwai, H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Nat.Chem.Biol., 9, 2013
|
|
3ZDE
 
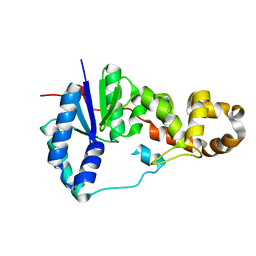 | | Potassium free structure of E. coli ExoIX | | Descriptor: | PROTEIN XNI | | Authors: | Hemsworth, G.R, Anstey-Gilbert, C.S, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZD8
 
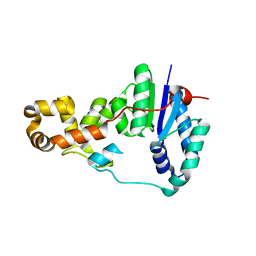 | | Potassium bound structure of E. coli ExoIX in P1 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
