7AQZ
 
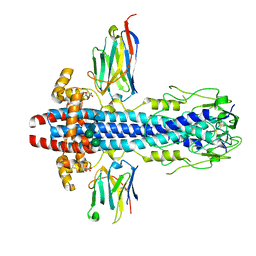 | | Co-Crystal Structure of Variant Surface Glycoprotein VSG2 in complex with Nanobody VSG2(NB14) | | Descriptor: | CITRIC ACID, Nanobody VSG2(NB14), SODIUM ION, ... | | Authors: | Stebbins, C.E, Hempelmann, A, VanStraaten, M. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome.
Cell Rep, 37, 2021
|
|
4BOR
 
 | |
4BRL
 
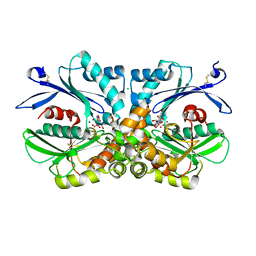 | | Legionella pneumophila NTPDase1 crystal form III (closed) in complex with transition state mimic guanosine 5'-phosphovanadate | | Descriptor: | CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GUANOSINE-5'-PHOSPHOVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
8H46
 
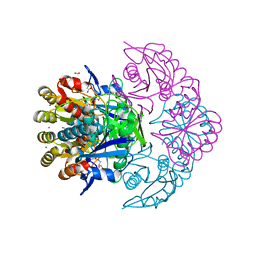 | | Blasnase-T13A/P55N with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, GLYCEROL, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H47
 
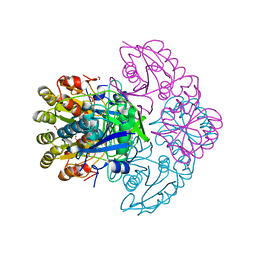 | | Blasnase-T13A/P55F | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
4BR9
 
 | | Legionella pneumophila NTPDase1 crystal form II, closed, apo | | Descriptor: | ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, GLYCEROL, SULFATE ION | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
8H4B
 
 | | Blasnase-T13A/M57P with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4E
 
 | | Blasnase-T13A/P55N with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
4BSO
 
 | | Crystal structure of R-spondin 1 (Fu1Fu2) - Native | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, R-SPONDIN-1 | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
8H4D
 
 | | Blasnase-T13A/M57N | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4J
 
 | | Crystal Structure of AzoR-FMN-Lyb24 complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase, ~{N}-(3-methylsulfanylphenyl)-4~{H}-cyclopenta[b]quinolin-9-amine | | Authors: | Huang, W. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of AzoR-FMN-Lyb24 complex
To Be Published
|
|
8H4M
 
 | | Crystal Structure of GTP-bound Irgb6_T95D mutant | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I
To Be Published
|
|
8H6B
 
 | | Crystal structure of AtHPPD complexed with YH20702 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-(1-methyl-1,2,3,4-tetrazol-5-yl)-1-pentyl-pyrrolo[2,3-b]pyridine-4-carboxamide | | Authors: | Yang, G.-F. | | Deposit date: | 2022-10-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Crystal structure of AtHPPD complexed with YH20702
To Be Published
|
|
4BWI
 
 | | Structure of the phytochrome Cph2 from Synechocystis sp. PCC6803 | | Descriptor: | FORMIC ACID, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Anders, K, Angerer, V, Widany, G.D, Mroginski, M.A, von Stetten, D, Essen, L.-O. | | Deposit date: | 2013-07-03 | | Release date: | 2013-10-30 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Cyanobacterial Phytochrome 2 Photosensor Implies a Tryptophan Switch for Phytochrome Signaling.
J.Biol.Chem., 288, 2013
|
|
8H8O
 
 | | Crystal structure of apo-R52W/E56W/R59W/E63W-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8H1R
 
 | | Crystal structure of LptDE-YifL complex | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
8H1S
 
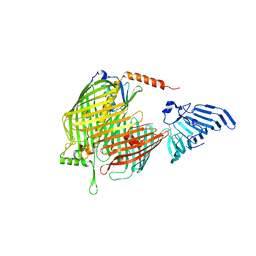 | | Crystal structure of apo-LptDE complex | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Luo, Q, Huang, Y. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Lipoprotein sorting to the cell surface via a crosstalk between the Lpt and Lol pathways during outer membrane biogenesis
To Be Published
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H1I
 
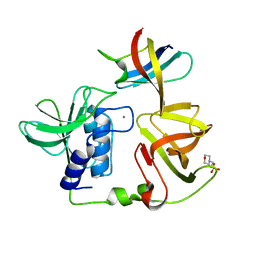 | | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cold shock-like protein CspC, ... | | Authors: | Padmanabhan, B, Gopinatha, K, Mandal, M, Saranya, G, Sudhagar, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PlyGRCS, a bacteriophage Endolysin in complex with Cold shock protein C
To Be Published
|
|
8H3H
 
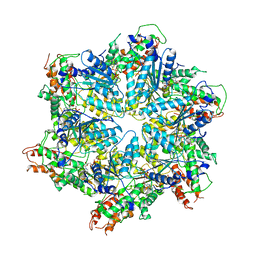 | | Human ATAD2 Walker B mutant, ATP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase family AAA domain-containing protein 2 | | Authors: | Cho, C, Song, J. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure of the human ATAD2 AAA+ histone chaperone reveals mechanism of regulation and inter-subunit communication.
Commun Biol, 6, 2023
|
|
8H44
 
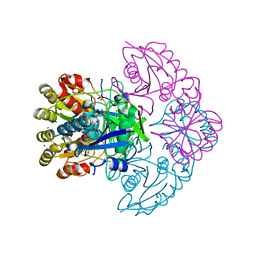 | | Blasnase-P55N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H45
 
 | | Blasnase-T13A/P55N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H48
 
 | | Blasnase-T13A/P55F with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
4C8A
 
 | | mouse ZNRF3 ectodomain crystal form II | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
