6E3V
 
 | |
6E04
 
 | | sperm whale myoglobin 1-nitrosopropane | | Descriptor: | 1-nitrosopropane, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
6E3W
 
 | |
6E3X
 
 | |
1C1Y
 
 | |
6EB1
 
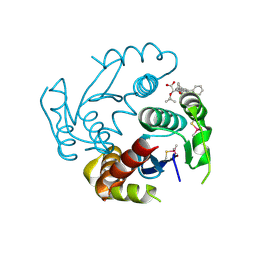 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid | | Descriptor: | (2S)-tert-butoxy[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-1-phenylisoquinolin-4-yl]acetic acid, Integrase | | Authors: | Lindenberger, J.J, Kobe, M, Kvaratskhelia, M. | | Deposit date: | 2018-08-03 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Isoquinoline Scaffold as a Novel Class of Allosteric HIV-1 Integrase Inhibitors.
ACS Med Chem Lett, 10, 2019
|
|
6EBS
 
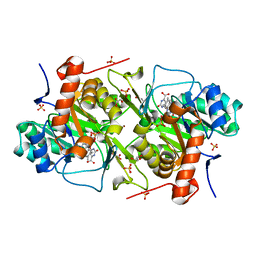 | | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate | | Descriptor: | Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Reis, R.A.G, Pinheiro, M.P, de Souza, A.L, Hunter, W.N, Nonato, M.C. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Leishmania major dihydroorotate dehydrogenase mutant H174A in complex with orotate
To Be Published
|
|
1CBI
 
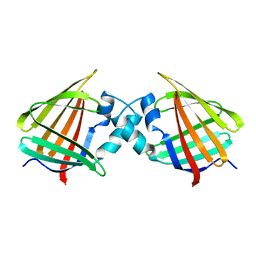 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN I | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN I | | Authors: | Thompson, J.R, Bratt, J.M, Banaszak, L.J. | | Deposit date: | 1995-07-12 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cellular retinoic acid binding protein I shows increased access to the binding cavity due to formation of an intermolecular beta-sheet.
J.Mol.Biol., 252, 1995
|
|
3DM0
 
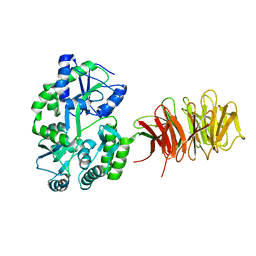 | | Maltose Binding Protein fusion with RACK1 from A. thaliana | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein fused with RACK1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ullah, H, Scappini, E.L, Moon, A.F, Williams, L.V, Armstrong, D.L, Pedersen, L.C. | | Deposit date: | 2008-06-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana.
Protein Sci., 17, 2008
|
|
6FCY
 
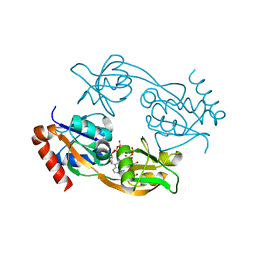 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP and ADP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
1ARB
 
 | |
6FB4
 
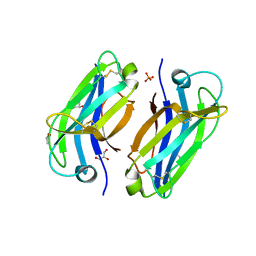 | | human KIBRA C2 domain mutant C771A | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein KIBRA | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.415631 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FJC
 
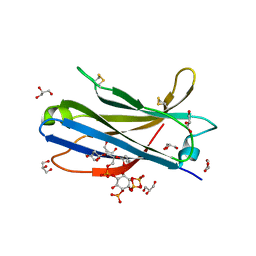 | | Human KIBRA C2 domain mutant C771A in complex with phosphatidylinositol 3,4,5-trisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, GLYCEROL, Protein KIBRA, ... | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
1B2A
 
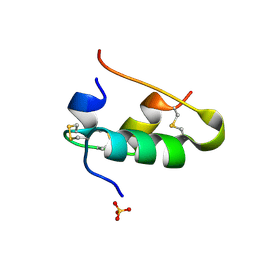 | |
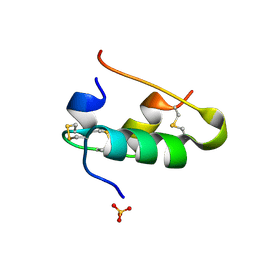
1B18
 
 | |
1B4Q
 
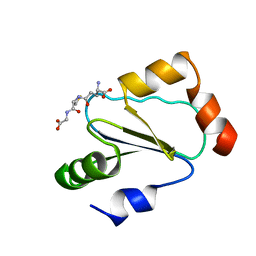 | | Solution structure of human thioltransferase complex with glutathione | | Descriptor: | GLUTATHIONE, PROTEIN (HUMAN THIOLTRANSFERASE) | | Authors: | Yang, Y, Jao, S.C, Nanduri, S, Starke, D.W, Mieyal, J.J, Qin, J. | | Deposit date: | 1998-12-25 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity.
Biochemistry, 37, 1998
|
|
1BCH
 
 | |
1B7Y
 
 | | PHENYLALANYL TRNA SYNTHETASE COMPLEXED WITH PHENYLALANINYL-ADENYLATE | | Descriptor: | ADENOSINE-5'-[PHENYLALANINOL-PHOSPHATE], MAGNESIUM ION, PROTEIN (PHENYLALANYL-TRNA SYNTHETASE) | | Authors: | Reshetnikova, L, Moor, N, Lavrik, O, Vassylyev, D.G. | | Deposit date: | 1999-01-26 | | Release date: | 2000-01-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of phenylalanyl-tRNA synthetase complexed with phenylalanine and a phenylalanyl-adenylate analogue.
J.Mol.Biol., 287, 1999
|
|
193L
 
 | | THE 1.33 A STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, SODIUM ION | | Authors: | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6GT8
 
 | |
1ALJ
 
 | | ALKALINE PHOSPHATASE MUTANT (H412N) | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ma, L, Tibbitts, T.T, Kantrowitz, E.R. | | Deposit date: | 1995-06-02 | | Release date: | 1995-11-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Escherichia coli alkaline phosphatase: X-ray structural studies of a mutant enzyme (His-412-->Asn) at one of the catalytically important zinc binding sites.
Protein Sci., 4, 1995
|
|
1AGH
 
 | |
1ABN
 
 | |
1ADU
 
 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
6ENK
 
 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
