4JJD
 
 | |
7R26
 
 | | PI3K delta in complex with SD5 | | Descriptor: | 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
6H8R
 
 | | CRYSTAL STRUCTURE OF THE HUMAN PROTEIN TYROSINE PHOSPHATASE PTPN5 (STEP) IN COMPLEX WITH COMPOUND 2 | | Descriptor: | 3-[(2~{S})-2-azanylpropyl]-5-(trifluoromethyl)phenol, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Hoerer, S, Fiegen, D, Schnapp, G. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Allosteric Activation of Striatal-Enriched Protein Tyrosine Phosphatase (STEP, PTPN5) by a Fragment-like Molecule.
J. Med. Chem., 62, 2019
|
|
7R2B
 
 | | PI3Kdelta in complex with an inhibitor | | Descriptor: | (4~{S})-3-[6-[2-azanyl-4-(trifluoromethyl)pyrimidin-5-yl]-2-morpholin-4-yl-pyrimidin-4-yl]-4-methyl-1,3-oxazolidin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Gutmann, S, Rummel, G, Shrestha, B. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
5OP2
 
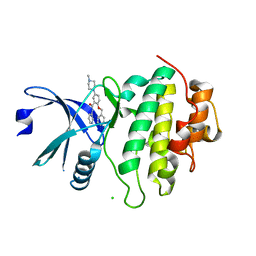 | | Structure of CHK1 10-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ8
 
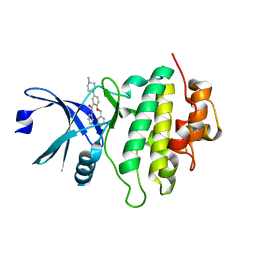 | | Structure of CHK1 12-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
8TE7
 
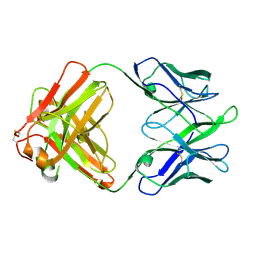 | | Structure of TRNM-f.01 | | Descriptor: | TRNM-f.01 Fab Heavy Chain, TRNM-f.01 Fab Light Chain | | Authors: | Bender, M.F, Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
1ZE3
 
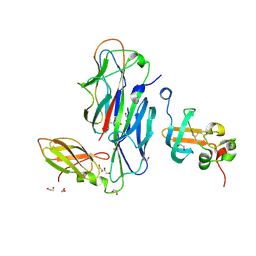 | | Crystal Structure of the Ternary Complex of FIMD (N-Terminal Domain) with FIMC and the Pilin Domain of FIMH | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein fimC, FimH protein, ... | | Authors: | Nishiyama, M, Horst, R, Eidam, O, Herrmann, T, Ignatov, O, Vetsch, M, Bettendorff, P, Jelesarov, I, Grutter, M.G, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-17 | | Release date: | 2005-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
8TJR
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-a.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody HERH-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TJS
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO GPZ6-a.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody GPZ6-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TNU
 
 | | Cryo-EM structure of TRNM-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TOP
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with antibody GPZ6-b.01 targeting the fusion peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP glycoprotein gp41, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
1W79
 
 | | Crystal structure of the DD-transpeptidase-carboxypeptidase from Actinomadura R39 | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, MAGNESIUM ION, SULFATE ION | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-31 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Actinomadura R39 DD-peptidase reveals new domains in penicillin-binding proteins.
J. Biol. Chem., 280, 2005
|
|
8TO7
 
 | | Cryo-EM structure of HERH-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HERH-b*01 heavy chain, ... | | Authors: | Roark, R.S, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL2
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL3
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-d.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP glycoprotein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL5
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-c.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
6I29
 
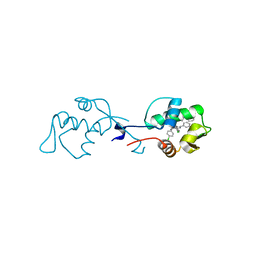 | | X-ray structure of the p53-MDM2 inhibitor NMI801 bound to HDM2 at 2.1A resolution | | Descriptor: | 6-chloranyl-3-[3-[(1~{S})-1-(4-chlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-~{N}-[2-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]pyridin-3-yl]-1~{H}-indole-2-carboxamide, Human E3 Ubiquitin-Protein Ligase MDM2 | | Authors: | Kallen, J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | p53 dynamics vary between tissues and are linked with radiation sensitivity
To be published
|
|
8TKC
 
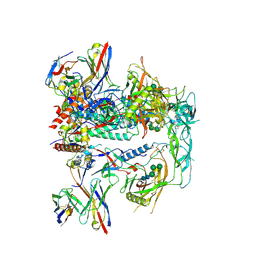 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-b.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-25 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TL4
 
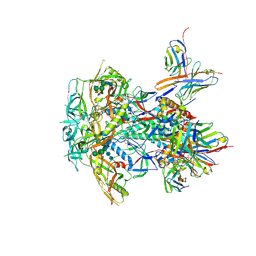 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO DJ85-e.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Pletnev, S, Hoyt, F, Fischer, E, Kwong, P. | | Deposit date: | 2023-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
5O2E
 
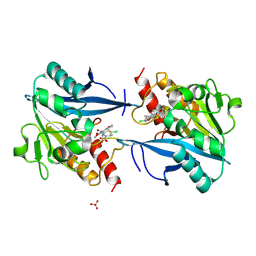 | | Crystal structure of NDM-1 in complex with hydrolyzed cefuroxime - new refinement | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
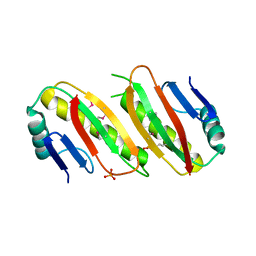
6QVP
 
 | |
1HYN
 
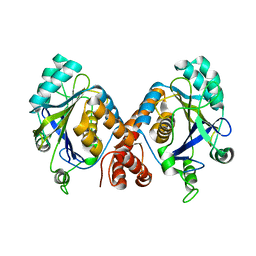 | |
8TTQ
 
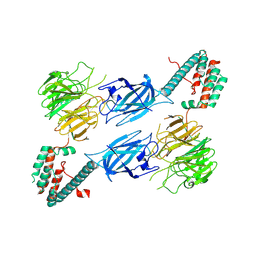 | |
7Y4K
 
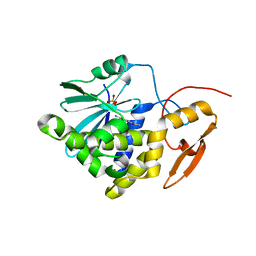 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-ornitine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-5-(phenylmethoxycarbonylamino)pentanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
