7MYU
 
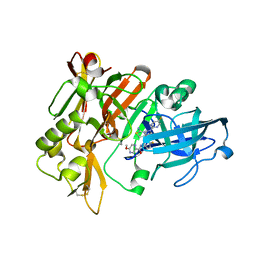 | | BACE-1 in complex with compound #22 | | Descriptor: | Beta-secretase 1, N-{3-[(4aR,7aS)-2-amino-6-(5-fluoropyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-4-fluorophenyl}-5-methoxypyrazine-2-carboxamide, SULFATE ION | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
7MYI
 
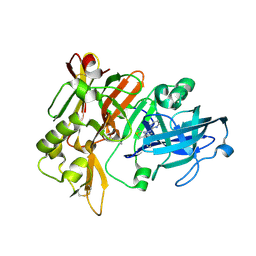 | | BACE-1 in complex with compound #6 | | Descriptor: | (4aR,7aR)-6-(pyrimidin-2-yl)-7a-(thiophen-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Hendle, J, Timm, D.E, Stout, S.L. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery and Early Clinical Development of LY3202626, a Low-Dose, CNS-Penetrant BACE Inhibitor.
J.Med.Chem., 64, 2021
|
|
2G0C
 
 | |
3MJ9
 
 | |
7Z8V
 
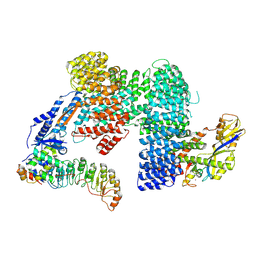 | | CAND1-SCF-SKP2 (SKP1deldel) CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8T
 
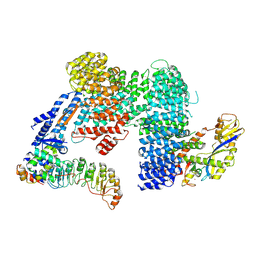 | | CAND1-SCF-SKP2 CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
4V48
 
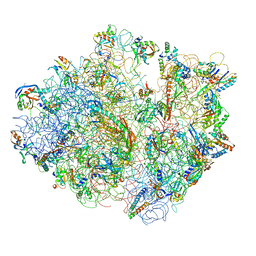 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
7ZBW
 
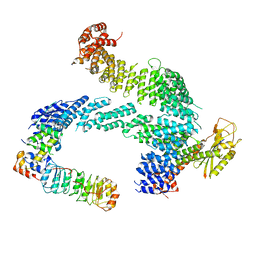 | | CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7N8N
 
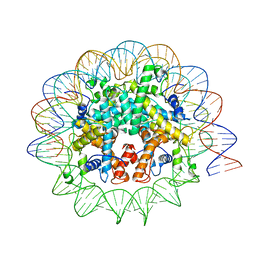 | | Melbournevirus nucleosome like particle | | Descriptor: | DNA (147-MER), Histone H2B-H2A doublet, Histone H4-H3 doublet | | Authors: | Liu, Y, Toner, C.M, Zhou, K, Bowerman, S, Luger, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Virus-encoded histone doublets are essential and form nucleosome-like structures.
Cell, 184, 2021
|
|
5I0S
 
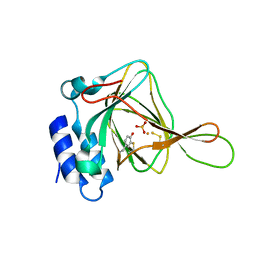 | | Thiosulfate bound Cysteine Dioxygenase at pH 6.2 | | Descriptor: | Cysteine dioxygenase type 1, FE (III) ION, THIOSULFATE | | Authors: | Kean, K.M, Driggers, C.M, Karplus, P.A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
7N9T
 
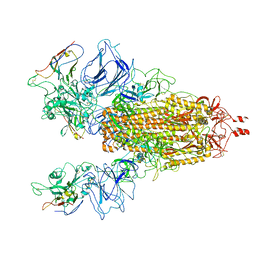 | |
4E4T
 
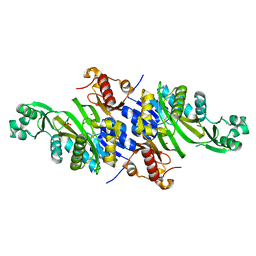 | |
3M7N
 
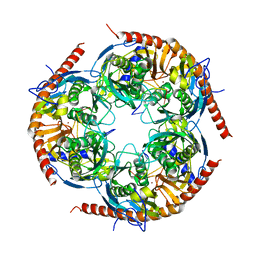 | |
3M86
 
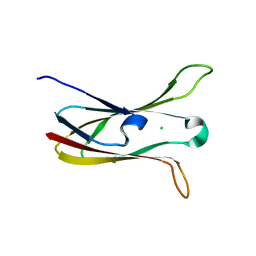 | | Crystal structure of the cysteine protease inhibitor, EhICP2, from Entamoeba histolytica | | Descriptor: | Amoebiasin-2, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Lara-Gonzalez, S, Casados-Vazquez, L.E, Brieba, L.G. | | Deposit date: | 2010-03-17 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the cysteine protease inhibitor 2 from Entamoeba histolytica: functional convergence of a common protein fold.
Gene, 471, 2011
|
|
2GHU
 
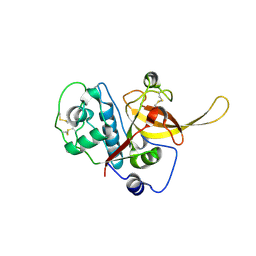 | | Crystal structure of falcipain-2 from Plasmodium falciparum | | Descriptor: | falcipain 2 | | Authors: | Hogg, T, Nagarajan, K, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2006-03-27 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Characterization of Falcipain-2, a Hemoglobinase from the Malarial Parasite Plasmodium falciparum.
J.Biol.Chem., 281, 2006
|
|
4UR8
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, KETO-DEOXY-D-GALACTARATE DEHYDRATASE | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2014-06-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and Function of a Decarboxylating Agrobacterium Tumefaciens Keto-Deoxy-D-Galactarate Dehydratase.
Biochemistry, 53, 2014
|
|
3MDX
 
 | |
4V4H
 
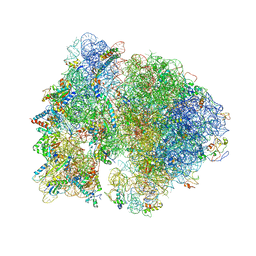 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with the antibiotic kasugamyin at 3.5A resolution. | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, ... | | Authors: | Schuwirth, B.S, Vila-Sanjurjo, A, Cate, J.H.D. | | Deposit date: | 2006-08-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structural analysis of kasugamycin inhibition of translation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5I25
 
 | |
4EQY
 
 | |
5IR5
 
 | | Crystal structure of wild-type bacterial lipoxygenase from Pseudomonas aeruginosa PA-LOX with space group P21212 at 1.9 A resolution | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, Arachidonate 15-lipoxygenase, FE (II) ION, ... | | Authors: | Kalms, J, Banthiya, S, Galemou Yoga, E, Kuhn, H, Scheerer, P. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional basis of phospholipid oxygenase activity of bacterial lipoxygenase from Pseudomonas aeruginosa.
Biochim.Biophys.Acta, 1861, 2016
|
|
3MNR
 
 | | Crystal Structure of Benzamide SNX-1321 bound to Hsp90 | | Descriptor: | 2-[(3,4,5-trimethoxyphenyl)amino]-4-(2,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Veal, J.M, Fadden, P, Huang, K.H, Rice, J, Hall, S.E, Haytstead, T.A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Chemoproteomics to Drug Discovery: Identification of a Clinical Candidate Targeting Hsp90.
Chem.Biol., 17, 2010
|
|
7Z6O
 
 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Varela, P.F, Charbonnier, J.B. | | Deposit date: | 2022-03-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
5I0R
 
 | | D-cysteine bound C93A mutant of Cysteine Dioxygenase at pH 8 | | Descriptor: | CHLORIDE ION, Cysteine dioxygenase type 1, D-CYSTEINE, ... | | Authors: | Kean, K.M, Driggers, C.M, Karplus, P.A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Insights into the Role of the Cys-Tyr Crosslink and Inhibitor Recognition by Mammalian Cysteine Dioxygenase.
J. Mol. Biol., 428, 2016
|
|
2GH0
 
 | | Growth factor/receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDNF family receptor alpha-3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q. | | Deposit date: | 2006-03-24 | | Release date: | 2006-06-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Artemin Complexed with Its Receptor GFRalpha3: Convergent Recognition of Glial Cell Line-Derived Neurotrophic Factors.
Structure, 14, 2006
|
|
