1JFA
 
 | |
1JFB
 
 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferric resting state at atomic resolution | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, nitric-oxide reductase cytochrome P450 55A1 | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JFC
 
 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferrous CO state at atomic resolution | | Descriptor: | CARBON MONOXIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JFD
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, SULFATE ION | | Authors: | Oganesyan, V, Avaeva, S.M, Huber, R, Harutyunyan, E.H. | | Deposit date: | 1997-05-31 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Escherichia coli inorganic pyrophosphatase complexed with SO4(2-). Ligand-induced molecular asymmetry.
FEBS Lett., 410, 1997
|
|
1JFF
 
 | | Refined structure of alpha-beta tubulin from zinc-induced sheets stabilized with taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lowe, J, Li, H, Downing, K.H, Nogales, E. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Refined structure of alpha beta-tubulin at 3.5 A resolution.
J.Mol.Biol., 313, 2001
|
|
1JFG
 
 | | TRICHODIENE SYNTHASE FROM FUSARIUM SPOROTRICHIOIDES COMPLEXED WITH DIPHOSPHATE | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Rynkiewicz, M.J, Cane, D.E, Christianson, D.W. | | Deposit date: | 2001-06-20 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JFH
 
 | |
1JFI
 
 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JFJ
 
 | | NMR SOLUTION STRUCTURE OF AN EF-HAND CALCIUM BINDING PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
1JFK
 
 | | MINIMUM ENERGY REPRESENTATIVE STRUCTURE OF A CALCIUM BOUND EF-HAND PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
1JFL
 
 | | CRYSTAL STRUCTURE DETERMINATION OF ASPARTATE RACEMASE FROM AN ARCHAEA | | Descriptor: | ASPARTATE RACEMASE | | Authors: | Liu, L.J, Iwata, K, Kita, A, Kawarabayasi, Y, Yohda, M, Miki, K. | | Deposit date: | 2001-06-21 | | Release date: | 2002-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartate racemase from Pyrococcus horikoshii OT3 and its implications for molecular mechanism of PLP-independent racemization.
J.Mol.Biol., 319, 2002
|
|
1JFM
 
 | |
1JFN
 
 | | SOLUTION STRUCTURE OF HUMAN APOLIPOPROTEIN(A) KRINGLE IV TYPE 6 | | Descriptor: | APOLIPOPROTEIN A, KIV-T6 | | Authors: | Maderegger, B, Bermel, W, Hrzenjak, A, Kostner, G.M, Sterk, H. | | Deposit date: | 2001-06-21 | | Release date: | 2002-06-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human apolipoprotein(a) kringle IV type 6.
Biochemistry, 41, 2002
|
|
1JFP
 
 | | Structure of bovine rhodopsin (dark adapted) | | Descriptor: | RETINAL, rhodopsin | | Authors: | Yeagle, P.L, Choi, G, Albert, A.D. | | Deposit date: | 2001-06-21 | | Release date: | 2001-10-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms.
Biochemistry, 40, 2001
|
|
1JFQ
 
 | | ANTIGEN-BINDING FRAGMENT OF THE MURINE ANTI-PHENYLARSONATE ANTIBODY 36-71, "FAB 36-71" | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF ANTI-PHENYLARSONATE ANTIBODY | | Authors: | Parhami-Seren, B, Viswanathan, M, Strong, R.K, Margolies, M.N. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of mutants of high-affinity and low-affinity
p-azophenylarsonate-specific antibodies generated by alanine
scanning of heavy chain
complementarity-determining region 2.
J.Immunol., 167, 2001
|
|
1JFR
 
 | |
1JFS
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PURF OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
1JFT
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PURF OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PHOSPHATE ION, ... | | Authors: | Huffman, J.L, Lu, F, Zalkin, H, Brennan, R.G. | | Deposit date: | 2001-06-21 | | Release date: | 2002-02-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Biochemistry, 41, 2002
|
|
1JFU
 
 | | CRYSTAL STRUCTURE OF THE SOLUBLE DOMAIN OF TLPA FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN TLPA | | Authors: | Capitani, G, Rossmann, R, Sargent, D.F, Gruetter, M.G, Richmond, T.J, Hennecke, H. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the soluble domain of a membrane-anchored thioredoxin-like protein from Bradyrhizobium japonicum reveals unusual properties.
J.Mol.Biol., 311, 2001
|
|
1JFV
 
 | | X-Ray Structure of oxidised C10S, C15A arsenate reductase from pI258 | | Descriptor: | PERCHLORATE ION, POTASSIUM ION, arsenate reductase | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2001-06-22 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty.
Nat.Struct.Biol., 8, 2001
|
|
1JFW
 
 | |
1JFX
 
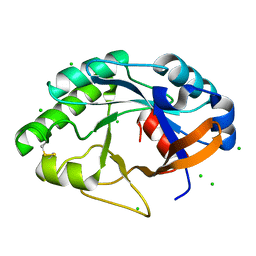 | | Crystal structure of the bacterial lysozyme from Streptomyces coelicolor at 1.65 A resolution | | Descriptor: | 1,4-beta-N-Acetylmuramidase M1, CHLORIDE ION | | Authors: | Rau, A, Hogg, T, Marquardt, R, Hilgenfeld, R. | | Deposit date: | 2001-06-22 | | Release date: | 2001-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new lysozyme fold. Crystal structure of the muramidase from Streptomyces coelicolor at 1.65 A resolution.
J.Biol.Chem., 276, 2001
|
|
1JFZ
 
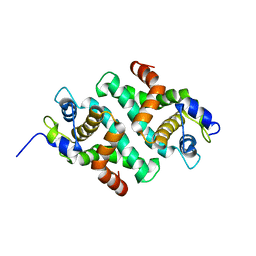 | |
1JG0
 
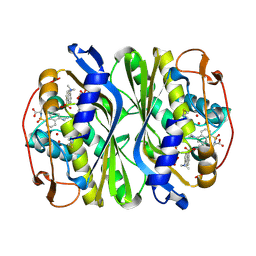 | | Crystal structure of Escherichia coli thymidylate synthase complexed with 2'-deoxyuridine-5'-monophosphate and N,O-didansyl-L-tyrosine | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N,O-DIDANSYL-L-TYROSINE, thymidylate synthase | | Authors: | Fritz, T.A, Tondi, D, Finer-Moore, J.S, Costi, M.P, Stroud, R.M. | | Deposit date: | 2001-06-22 | | Release date: | 2002-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Predicting and harnessing protein flexibility in the design of species-specific inhibitors of thymidylate synthase.
Chem.Biol., 8, 2001
|
|
1JG1
 
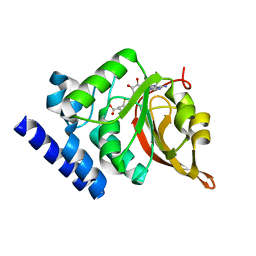 | | Crystal Structure of L-isoaspartyl (D-aspartyl) O-methyltransferase with S-ADENOSYL-L-HOMOCYSTEINE | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein-L-isoaspartate O-methyltransferase | | Authors: | Griffith, S.C, Sawaya, M.R, Boutz, D, Thapar, N, Katz, J, Clarke, S, Yeates, T.O. | | Deposit date: | 2001-06-22 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its L-isoaspartyl peptide substrate.
J.Mol.Biol., 313, 2001
|
|
