6RS7
 
 | | X-ray crystal structure of LsAA9B (deglycosylated form) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AA9, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
7Q3X
 
 | |
6JI1
 
 | |
7YMB
 
 | |
4J9C
 
 | |
1DTO
 
 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
6JRG
 
 | | Crystal structure of ZmMoc1 H253A mutant in complex with Holliday junction | | Descriptor: | DNA (32-MER), DNA (33-MER), MAGNESIUM ION, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
7PXX
 
 | | The crystal structure of Leishmania major Pteridine Reductase 1 in complex with substrate folic acid | | Descriptor: | DI(HYDROXYETHYL)ETHER, FOLIC ACID, GLYCEROL, ... | | Authors: | Di Pisa, F, Dello Iacono, L, Mangani, S. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the ternary complex of Leishmania major pteridine reductase 1 with the cofactor NADP + /NADPH and the substrate folic acid.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6RS8
 
 | | X-ray crystal structure of LsAA9B (transition metals soak) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frandsen, K.E.H, Tovborg, M, Poulsen, J.C.N, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2019-05-21 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insights into an unusual Auxiliary Activity 9 family member lacking the histidine brace motif of lytic polysaccharide monooxygenases.
J.Biol.Chem., 294, 2019
|
|
5NRE
 
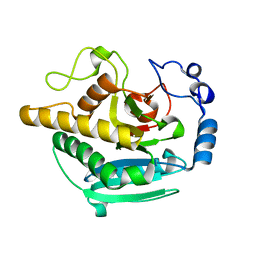 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | Descriptor: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6JKD
 
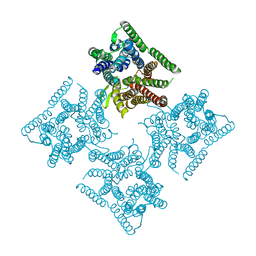 | | Crystal structure of tetrameric PepTSo2 in I4 space group | | Descriptor: | Proton:oligopeptide symporter POT family | | Authors: | Nagamura, R, Fukuda, M, Ishitani, R, Nureki, O. | | Deposit date: | 2019-02-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis for oligomerization of the prokaryotic peptide transporter PepTSo2.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4J9G
 
 | |
1QNW
 
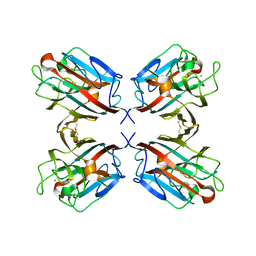 | | lectin II from Ulex europaeus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-10-22 | | Release date: | 1999-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
5NRD
 
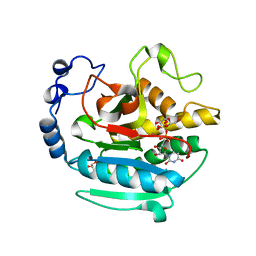 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-2 | | Descriptor: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8SUX
 
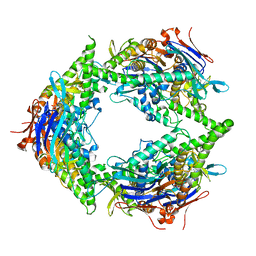 | | Structure of E. coli PtuA hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7Z8O
 
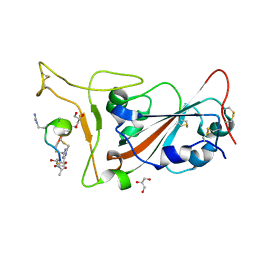 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8SXI
 
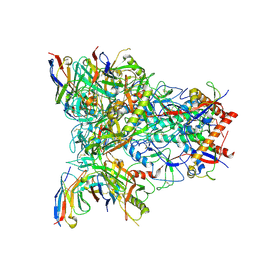 | | CH505 Disulfide Stapled SOSIP Bound to b12 Fab | | Descriptor: | Envelope glycoprotein gp160, HIV-1 gp41, b12 Heavy Chain, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
8SXJ
 
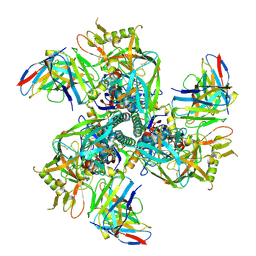 | | CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab | | Descriptor: | CH235.12 Heavy Chain, CH235.12 Light Chain, Envelope glycoprotein gp160, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
5NR9
 
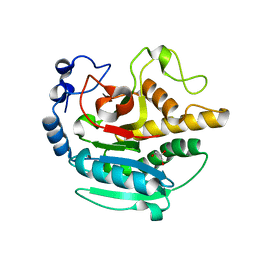 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (alpha-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - Unliganded alpha-3GalT | | Descriptor: | GLYCEROL, N-acetyllactosaminide alpha-1,3-galactosyltransferase | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6RPU
 
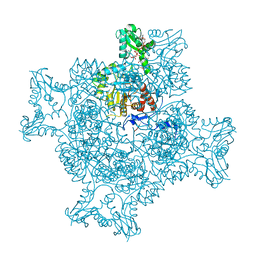 | | Structure of the ternary complex of the IMPDH enzyme from Ashbya gossypii bound to the dinucleoside polyphosphate Ap5G and GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates.
J.Biol.Chem., 294, 2019
|
|
5NBK
 
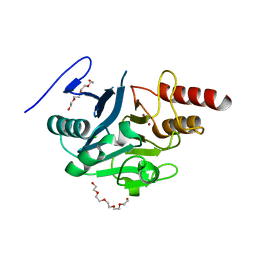 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5N0I
 
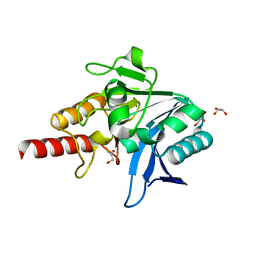 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1QOO
 
 | | lectin UEA-II complexed with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHITIN BINDING LECTIN, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-15 | | Release date: | 1999-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
1WT6
 
 | | Coiled-Coil domain of DMPK | | Descriptor: | Myotonin-protein kinase | | Authors: | Garcia, P, Marino, M, Mayans, O. | | Deposit date: | 2004-11-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into the self-assembly mechanism of dystrophia myotonica kinase
Faseb J., 20, 2006
|
|
6JER
 
 | | Apo crystal structure of class I type a peptide deformylase from Acinetobacter baumannii | | Descriptor: | Peptide deformylase, ZINC ION | | Authors: | Ho, T.H, Lee, I.H, Kang, L.W. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, crystallization, and preliminary X-ray crystallographic analysis of peptide deformylase from Acinetobacter baumanii
Biodesign, 5, 2017
|
|
