3AXB
 
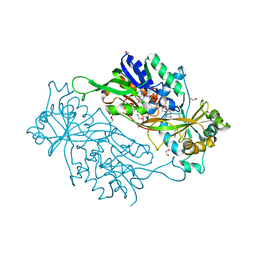 | | Structure of a dye-linked L-proline dehydrogenase from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, PROLINE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K, Hara, Y. | | Deposit date: | 2011-04-01 | | Release date: | 2012-04-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
3AYT
 
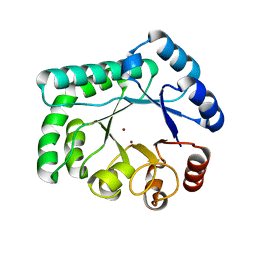 | | TTHB071 protein from Thermus thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
3AXX
 
 | |
3AZ1
 
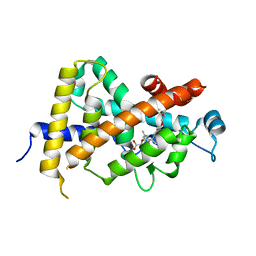 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | Vitamin D3 receptor, {4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}acetic acid | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AY5
 
 | |
3AZ2
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | 5-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}-3-methylphenyl)pentan-3-yl]-2-methylphenoxy}pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AY3
 
 | |
3AZ3
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (4S)-4-hydroxy-5-[4-(3-{4-[(3S)-3-hydroxy-4,4-dimethylpentyl]-3-methylphenyl}pentan-3-yl)-2-methylphenoxy]pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Novel nonsecosteroidal vitamin D(3) carboxylic acid analogs for osteoporosis, and SAR analysis.
Bioorg.Med.Chem., 19, 2011
|
|
3AYV
 
 | | TTHB071 protein from Thermus thermophilus HB8 soaking with ZnCl2 | | Descriptor: | Putative uncharacterized protein TTHB071, ZINC ION | | Authors: | Nakane, S, Wakamatsu, T, Fukui, K, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vivo, in vitro, and x-ray crystallographic analyses suggest the involvement of an uncharacterized triose-phosphate isomerase (TIM) barrel protein in protection against oxidative stress
J.Biol.Chem., 286, 2011
|
|
3B0L
 
 | | M175G mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B0V
 
 | | tRNA-dihydrouridine synthase from Thermus thermophilus in complex with tRNA | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-14 | | Release date: | 2011-12-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B0Y
 
 | | K263D mutant of PolX from Thermus thermophilus HB8 complexed with Ca-dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakane, S, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-17 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA.
J.Mol.Biol., 417, 2012
|
|
3B26
 
 | | Hsp90 alpha N-terminal domain in complex with an inhibitor Ro1127850 | | Descriptor: | 4-(1H,3H-benzo[de]isochromen-6-yl)-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-07-21 | | Release date: | 2011-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lead generation of heat shock protein 90 inhibitors by a combination of fragment-based approach, virtual screening, and structure-based drug design
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3B8U
 
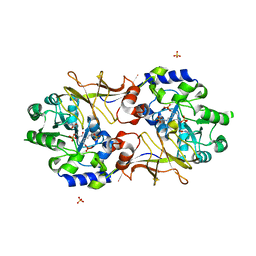 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3BE2
 
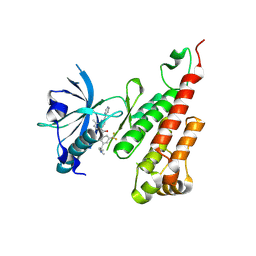 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzamide inhibitor | | Descriptor: | N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Gu, Y, Rose, P, Zhao, H. | | Deposit date: | 2007-11-16 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Naphthamides as novel and potent vascular endothelial growth factor receptor tyrosine kinase inhibitors: design, synthesis, and evaluation.
J.Med.Chem., 51, 2008
|
|
3BIX
 
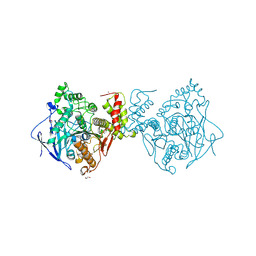 | | Crystal structure of the extracellular esterase domain of Neuroligin-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
3B0J
 
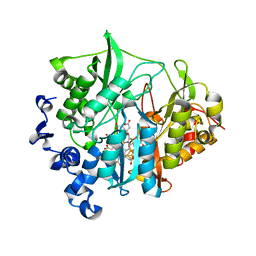 | | M175E mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B2J
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3B6Q
 
 | |
3B6W
 
 | |
3B0H
 
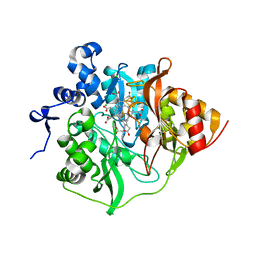 | | Assimilatory nitrite reductase (Nii4) from tobbaco root | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B2I
 
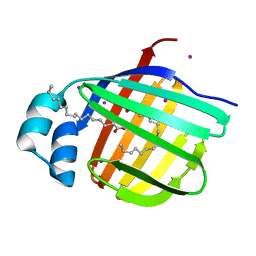 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3BFU
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
2ZWB
 
 | | Neutron crystal structure of wild type human lysozyme in D2O | | Descriptor: | Lysozyme C | | Authors: | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme
To be Published
|
|
2ZSY
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [750 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
