7Y8A
 
 | | Cryo-EM structure of cryptophyte photosystem I | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Zhao, L.S, Zhang, Y.Z, Liu, L.N, Li, K. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis and evolution of the photosystem I-light-harvesting supercomplex of cryptophyte algae.
Plant Cell, 35, 2023
|
|
4EI4
 
 | | JAK1 kinase (JH1 domain) in complex with compound 20 | | Descriptor: | (1R,3R)-3-(2-methylimidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(8H)-yl)cyclohexanol, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
7XRZ
 
 | | Crystal structure of BRIL and SRP2070_Fab complex | | Descriptor: | IGG HEAVY CHAIN, IGG LIGHT CHAIN, Soluble cytochrome b562 | | Authors: | Suzuki, M, Miyagi, H, Yasunaga, M, Asada, H, Iwata, S, Saito, J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into an anti-BRIL Fab as a G-protein-coupled receptor crystallization chaperone.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3KK9
 
 | | CaMKII Substrate Complex B | | Descriptor: | Calcium/calmodulin dependent protein kinase II | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-04 | | Release date: | 2010-02-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4IW2
 
 | | HSA-glucose complex | | Descriptor: | D-glucose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Luo, Z, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
4F09
 
 | |
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
7Y9I
 
 | | Complex structure of AtYchF1 with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Co-crystalization reveals the interaction between AtYchF1 and ppGpp.
Front Mol Biosci, 9, 2022
|
|
4ITQ
 
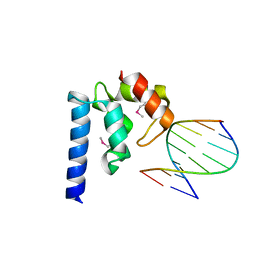 | | Crystal structure of hypothetical protein SCO1480 bound to DNA | | Descriptor: | 5'-D(P*CP*CP*GP*CP*GP*CP*GP*C)-3', 5'-D(P*GP*CP*GP*CP*GP*CP*GP*G)-3', Putative uncharacterized protein SCO1480 | | Authors: | Guarne, A, Nanji, T, Gloyd, M, Swiercz, J.P, Elliot, M.A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-27 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel nucleoid-associated protein specific to the actinobacteria.
Nucleic Acids Res., 41, 2013
|
|
1NRP
 
 | |
2I4N
 
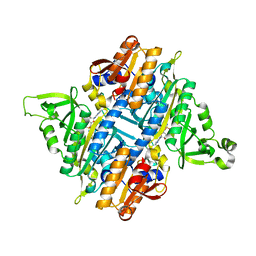 | | Rhodopseudomonas palustris prolyl-tRNA synthetase in complex with CysAMS | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Proline-tRNA ligase | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a cis-Editing Domain.
Structure, 14, 2006
|
|
2I4M
 
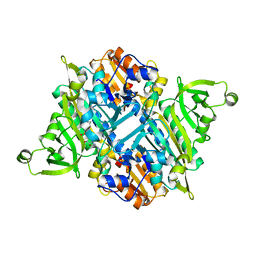 | | Rhodopseudomonas palustris prolyl-tRNA synthetase in complex with ProAMS | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, Proline-tRNA ligase | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a cis-Editing Domain.
Structure, 14, 2006
|
|
4IW1
 
 | | HSA-fructose complex | | Descriptor: | D-fructose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
7Z71
 
 | | Crystal structure of p63 DBD in complex with darpin C14 | | Descriptor: | Darpin C14, Isoform 4 of Tumor protein 63, ZINC ION | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z73
 
 | | Crystal structure of p63 tetramerization domain in complex with darpin 8F1 | | Descriptor: | Darpin 8F1, Isoform 2 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7Z2E
 
 | |
7YTA
 
 | |
4NZ4
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with 2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE (NDG) and zinc ion | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4O46
 
 | | 14-3-3-gamma in complex with influenza NS1 C-terminal tail phosphorylated at S228 | | Descriptor: | 14-3-3 protein gamma, Nonstructural protein 1, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Liu, Y, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
4NW2
 
 | | Tandem chromodomains of human CHD1 in complex with Influenza virus NS1 C-terminal tail trimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Tempel, W, Xu, C, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
4NZ3
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7Z8E
 
 | | Crystal structure of the substrate-binding protein YejA from S. meliloti in complex with peptide fragment | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, GLY-SER-ASP-VAL-ALA, ... | | Authors: | Morera, S, Vigouroux, V, Travin, D.Y. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual-Uptake Mode of the Antibiotic Phazolicin Prevents Resistance Acquisition by Gram-Negative Bacteria.
Mbio, 14, 2023
|
|
4ND2
 
 | |
4ND1
 
 | |
7YPP
 
 | | Structural basis of a superoxide dismutase from a tardigrade, Ramazzottius varieornatus strain YOKOZUNA-1. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Sim, K.-S, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a superoxide dismutase from a tardigrade: Ramazzottius varieornatus strain YOKOZUNA-1.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
