7B8F
 
 | | Notum-Fragment 154 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8O
 
 | | Notum-Fragment 199 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B86
 
 | | Notum-Fragment067 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(1~{H}-pyrazol-3-yl)phenoxy]pyrimidine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8L
 
 | | Notum-Fragment 174 | | Descriptor: | 1,2-ETHANEDIOL, 2,4-bis(fluoranyl)-6-(1~{H}-pyrazol-3-yl)phenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
9CWS
 
 | | Bufavirus 1 at pH 2.6 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
7B8K
 
 | | Notum-Fragment 173 | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-benzimidazol-2-ylcyanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
6HDN
 
 | |
8ZET
 
 | | Tp-PSI-FCPI-S in Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z, Shen, J.R, Wang, W. | | Deposit date: | 2024-05-06 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of PSI-FCPI from Thalassiosira pseudonana grown under high light provide evidence for convergent evolution and light-adaptive strategies in diatom FCPIs.
J Integr Plant Biol, 67, 2025
|
|
5XKM
 
 | | Crystal structure of human phosphodiesterase 2A in complex with 6-methyl-N-(1-(4-(trifluoromethoxy)phenyl)propyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | 6-methyl-N-[(1R)-1-[4-(trifluoromethyloxy)phenyl]propyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Kondo, M, Snell, G, Lane, W. | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of an Orally Bioavailable, Brain-Penetrating, in Vivo Active Phosphodiesterase 2A Inhibitor Lead Series for the Treatment of Cognitive Disorders.
J. Med. Chem., 60, 2017
|
|
7RTB
 
 | | Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Johnson, R.M, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2021-08-12 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
8RDC
 
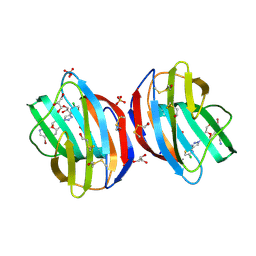 | | Galectin-1 in complex with thiogalactoside derivative | | Descriptor: | 5-bromanyl-3-[(2~{R},3~{R},4~{S},5~{R},6~{R})-4-[4-(4-chloranyl-1,3-thiazol-2-yl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3-methoxy-5-oxidanyl-oxan-2-yl]sulfanyl-pyridine-2-carbonitrile, GLYCEROL, Galectin-1, ... | | Authors: | Hakansson, M, Diehl, C, Peterson, K, Zetterberg, F, Nilsson, U. | | Deposit date: | 2023-12-07 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Galectin-1 in complex with thiogalactoside derivative
To Be Published
|
|
9D6E
 
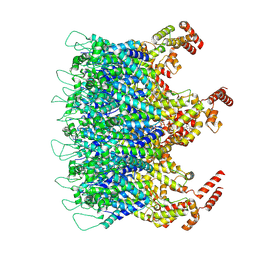 | | Gag CA-SP1 immature lattice bound with Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
7S02
 
 | |
6OPQ
 
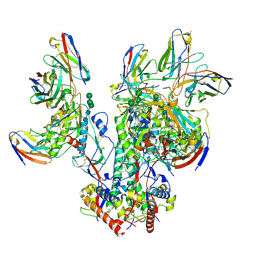 | | CD4- and 17-bound HIV-1 Env B41 SOSIP frozen with LMNG | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
9CWV
 
 | |
9D88
 
 | |
5NBU
 
 | |
9D6C
 
 | | Gag CA-SP1 immature lattice bound with Lenacapavir and Bevirimat from enveloped virus like particles | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wu, C, Meuser, M.E, Xiong, Y. | | Deposit date: | 2024-08-14 | | Release date: | 2024-10-30 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural insights into inhibitor mechanisms on immature HIV-1 Gag lattice revealed by high-resolution in situ single-particle cryo-EM.
Biorxiv, 2024
|
|
9FRE
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with THIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Last modified: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Cryo-EM structures of rho 1 GABA A receptors with antagonist and agonist drugs.
Nat Commun, 16, 2025
|
|
8WDQ
 
 | | Crystal Structure of Pseudomonas aeruginosa SuhB in complex with D-myo-inositol-1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadav, V.K, Maji, S, Shukla, M, Bhattacharyya, S. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Pseudomonas aeruginosa SuhB in complex with D-myo-inositol-1-phosphate
To Be Published
|
|
5NE5
 
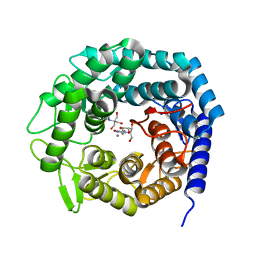 | | Crystal structure of family 47 alpha-1,2-mannosidase from Caulobacter K31 strain in complex with kifunensine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, KIFUNENSINE, ... | | Authors: | Males, A, Davies, G.J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Conformational Analysis of the Mannosidase Inhibitor Kifunensine: A Quantum Mechanical and Structural Approach.
Chembiochem, 18, 2017
|
|
6X4M
 
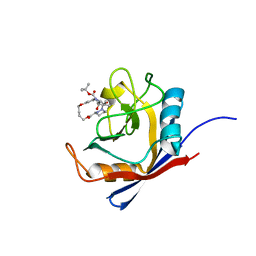 | | Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate (compound 3) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, tert-butyl [(2S)-1-{[(3S,17S)-2,16-dioxo-10,15-dioxa-1,21-diazatricyclo[15.3.1.1~5,9~]docosa-5(22),6,8-trien-3-yl]amino}-3-methyl-1-oxobutan-2-yl]carbamate | | Authors: | Appleby, T.C, Paulsen, J.L, Schmitz, U, Shivakumar, D. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of Free Energy Calculations for the Prioritization of Macrocycle Synthesis.
J.Chem.Inf.Model., 60, 2020
|
|
7RXS
 
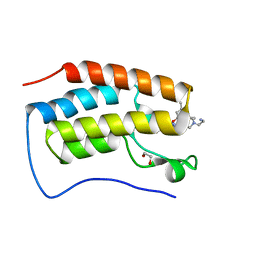 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7R9C
 
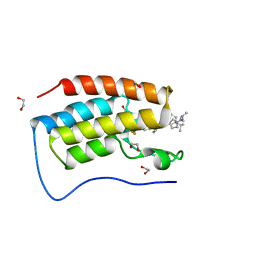 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
8OMT
 
 | | X-ray structure of lysozyme obtained upon reaction with [VIVO(empp)2] (Structure C) | | Descriptor: | 1-methyl-2-ethyl-3-hydroxy-4(1H)-pyridinone)V(IV)O4, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C, ... | | Authors: | Paolillo, M, Merlino, A, Ferraro, G. | | Deposit date: | 2023-03-31 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Implications of Protein Interaction in the Speciation of Potential V IV O-Pyridinone Drugs.
Inorg.Chem., 62, 2023
|
|
