5C2L
 
 | |
5BYT
 
 | |
5C1R
 
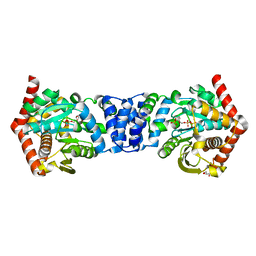 | | Stereoisomer of PRPP bound in the active site of Mycobacterium tuberculosis anthranilate phosphoribosyl (AnPRT; trpD) | | Descriptor: | 5-O-[(R)-hydroxy(phosphonooxy)phosphoryl]-1-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, GLYCEROL, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-06-15 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Binding and mimicking of the phosphate-rich substrate, PRPP.
To Be Published
|
|
5C7S
 
 | |
5BO2
 
 | |
8R1Z
 
 | |
8R1U
 
 | |
3WO1
 
 | | Crystal structure of Trp332Ala mutant YwfE, an L-amino acid ligase, with bound ADP-Mg-Ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, Alanine-anticapsin ligase BacD, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
4QKS
 
 | | Crystal Structure of 6xTrp/PV2: de novo designed beta-trefoil architecture with symmetric primary structure (L22W/L44W/L64W/L85W/L108W/L132W his Primitive Version 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DE NOVO PROTEIN 6XTRP/PV2, SULFATE ION | | Authors: | Longo, L.M, Tenorio, C.A, Blaber, M. | | Deposit date: | 2014-06-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A single aromatic core mutation converts a designed "primitive" protein from halophile to mesophile folding.
Protein Sci., 24, 2015
|
|
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
4AAJ
 
 | | Structure of N-(5'-phosphoribosyl)anthranilate isomerase from Pyrococcus furiosus | | Descriptor: | N-(5'-PHOSPHORIBOSYL)ANTHRANILATE ISOMERASE, SULFATE ION | | Authors: | Repo, H, Oeemig, J.S, Djupsjobacka, J.B, Iwai, H, Heikinheimo, P. | | Deposit date: | 2011-12-04 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Monomeric Tim-Barrel Structure from Pyrococcus Furiosus is Optimized for Extreme Temperatures
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2WUC
 
 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp and Ac-KQLR-chloromethylketone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACE-KQLR-CHLOROMETHYLKETONE INHIBITOR, FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
5Z4W
 
 | | Crystal structure of signalling protein from buffalo (SPB-40) with an altered conformation of Trp78 at 1.79 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Singh, P.K, Chaudhary, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-01-15 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A glycoprotein from mammary gland secreted during involution promotes apoptosis: Structural and biological studies.
Arch. Biochem. Biophys., 644, 2018
|
|
1B80
 
 | | REC. LIGNIN PEROXIDASE H8 OXIDATIVELY PROCESSED | | Descriptor: | CALCIUM ION, PROTEIN (RECOMBINANT LIGNIN PEROXIDASE H8), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Blodig, W, Smith, A.T, Doyle, W.A, Piontek, K. | | Deposit date: | 1999-02-03 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism.
J.Mol.Biol., 305, 2001
|
|
2WUB
 
 | | Crystal structure of HGFA in complex with the allosteric non- inhibitory antibody Fab40.deltaTrp | | Descriptor: | FAB FRAGMENT FAB40.DELTATRP HEAVY CHAIN, FAB FRAGMENT FAB40.DELTATRP LIGHT CHAIN, HEPATOCYTE GROWTH FACTOR ACTIVATOR LONG CHAIN, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the Allosteric Mechanism of Serine Protease Inhibition by an Antibody
Structure, 17, 2009
|
|
4KPR
 
 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
5Z05
 
 | | Crystal structure of signalling protein from buffalo (SPB-40) with an acetone induced conformation of Trp78 at 1.49 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONE, ... | | Authors: | Singh, P.K, Chaudhary, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A glycoprotein from mammary gland secreted during involution promotes apoptosis: Structural and biological studies
Arch. Biochem. Biophys., 644, 2018
|
|
1B82
 
 | | PRISTINE RECOMB. LIGNIN PEROXIDASE H8 | | Descriptor: | CALCIUM ION, PROTEIN (LIGNIN PEROXIDASE), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Blodig, W, Doyle, W.A, Smith, A.T, Piontek, K. | | Deposit date: | 1999-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism.
J.Mol.Biol., 305, 2001
|
|
1O17
 
 | |
2GVQ
 
 | | Anthranilate phosphoribosyl-transferase (TRPD) from S. solfataricus in complex with anthranilate | | Descriptor: | 2-AMINOBENZOIC ACID, Anthranilate phosphoribosyltransferase | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2006-05-03 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
2KJ4
 
 | | Solution structure of the complex of VEK-30 and plasminogen kringle 2 | | Descriptor: | VEK-30, plasminogen | | Authors: | Wang, M, Zajicek, J, Prorok, M, Castellin, F.J. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the complex of VEK-30 and plasminogen kringle 2.
J.Struct.Biol., 169, 2010
|
|
1I5K
 
 | | STRUCTURE AND BINDING DETERMINANTS OF THE RECOMBINANT KRINGLE-2 DOMAIN OF HUMAN PLASMINOGEN TO AN INTERNAL PEPTIDE FROM A GROUP A STREPTOCOCCAL SURFACE PROTEIN | | Descriptor: | M PROTEIN, PLASMINOGEN | | Authors: | Rios-Steiner, J.L, Schenone, M, Mochalkin, I, Tulinsky, A, Castellino, F.J. | | Deposit date: | 2001-02-27 | | Release date: | 2001-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein.
J.Mol.Biol., 308, 2001
|
|
1QA5
 
 | |
1F1W
 
 | | SRC SH2 THREF1TRP MUTANT COMPLEXED WITH THE PHOSPHOPEPTIDE S(PTR)VNVQN | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC, S(PTR)VNVQN PHOSPHOPEPTIDE | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-20 | | Release date: | 2000-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
1F2F
 
 | | SRC SH2 THREF1TRP MUTANT | | Descriptor: | PHOSPHATE ION, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Kimber, M.S, Nachman, J, Cunningham, A.M, Gish, G.D, Pawson, T, Pai, E.F. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for specificity switching of the Src SH2 domain.
Mol.Cell, 5, 2000
|
|
