6QEB
 
 | |
5KJG
 
 | | Connexin 26 G12R mutant NMR structure | | Descriptor: | Gap junction beta-2 protein | | Authors: | Dowd, T.L, Bargiello, T.A. | | Deposit date: | 2016-06-19 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Structural studies of N-terminal mutants of Connexin 26 and Connexin 32 using (1)H NMR spectroscopy.
Arch.Biochem.Biophys., 608, 2016
|
|
7M0G
 
 | |
7M3U
 
 | |
1W6B
 
 | | Solution NMR Structure of a Long Neurotoxin from the Venom of the Asian Cobra, 20 Structures | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Talebzadeh-Farooji, M, Amininasab, M, Elmi, M.M, Naderi-Manesh, H, Sarbolouki, M.N. | | Deposit date: | 2004-08-17 | | Release date: | 2004-12-22 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of long neurotoxin NTX-1 from the venom of Naja naja oxiana by 2D-NMR spectroscopy.
Eur. J. Biochem., 271, 2004
|
|
7LOI
 
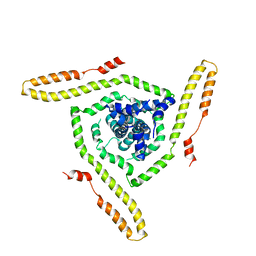 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
1VHP
 
 | | VH-P8, NMR | | Descriptor: | VH-P8 | | Authors: | Riechmann, L. | | Deposit date: | 1996-04-16 | | Release date: | 1996-11-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Rearrangement of the former VL interface in the solution structure of a camelised, single antibody VH domain.
J.Mol.Biol., 259, 1996
|
|
1M8M
 
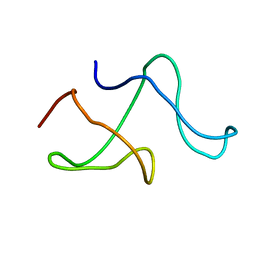 | | SOLID-STATE MAS NMR STRUCTURE OF THE A-SPECTRIN SH3 DOMAIN | | Descriptor: | SPECTRIN ALPHA CHAIN, BRAIN | | Authors: | Castellani, F, Van Rossum, B, Diehl, A, Schubert, M, Rehbein, K, Oschkinat, H. | | Deposit date: | 2002-07-25 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy
Nature, 420, 2002
|
|
1MIC
 
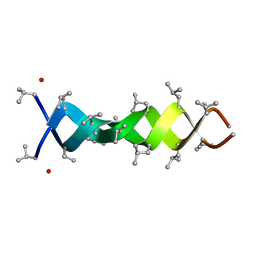 | |
8H1D
 
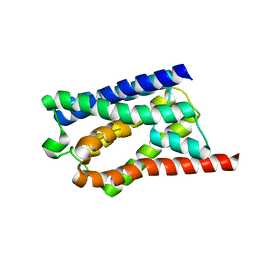 | | Solid-state NMR Structure of Aquaporin Z in its Native Cellular Membranes | | Descriptor: | Aquaporin Z | | Authors: | Xie, H, Zhao, Y, Zhao, W, Chen, Y, Liu, M, Yang, J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure determination of a membrane protein in E. coli cellular inner membrane.
Sci Adv, 9, 2023
|
|
8GQO
 
 | |
4TGF
 
 | | SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA | | Descriptor: | DES-VAL-1,VAL-2,TRANSFORMING GROWTH FACTOR, ALPHA | | Authors: | Kline, T.P, Brown, F.K, Brown, S.C, Jeffs, P.W, Kopple, K.D, Mueller, L. | | Deposit date: | 1990-06-13 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human transforming growth factor alpha derived from 1H NMR data.
Biochemistry, 29, 1990
|
|
1MR0
 
 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
5GIW
 
 | | Solution NMR structure of Humanin containing a D-isomerized serine residue | | Descriptor: | Humanin | | Authors: | Furuita, K, Sugiki, T, Alsanousi, N, Fujiwara, T, Kojima, C. | | Deposit date: | 2016-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and inhibitory effect against amyloid-beta fibrillation of Humanin containing a d-isomerized serine residue
Biochem.Biophys.Res.Commun., 477, 2016
|
|
1VSQ
 
 | |
1MWN
 
 | | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12 | | Descriptor: | CALCIUM ION, F-actin capping protein alpha-1 subunit, S-100 protein, ... | | Authors: | Inman, K.G, Yang, R, Rustandi, R.R, Miller, K.E, Baldisseri, D.M, Weber, D.J. | | Deposit date: | 2002-09-30 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
J.Mol.Biol., 324, 2002
|
|
7OTD
 
 | | Oxytocin NMR solution structure | | Descriptor: | AMINO GROUP, COPPER (II) ION, UNK-TYR-ILE-GLN-ASN-CYS-PRO-LEU-GLY | | Authors: | Shalev, D.E, Alshanski, I, Yitzchaik, S, Hurevich, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-13 | | Method: | SOLUTION NMR | | Cite: | Determining the structure and binding mechanism of oxytocin-Cu 2+ complex using paramagnetic relaxation enhancement NMR analysis.
J.Biol.Inorg.Chem., 26, 2021
|
|
1MM4
 
 | | Solution NMR structure of the outer membrane enzyme PagP in DPC micelles | | Descriptor: | CrcA protein | | Authors: | Hwang, P.M, Choy, W.-Y, Lo, E.I, Chen, L, Forman-Kay, J.D, Raetz, C.R.H, Prive, G.G, Bishop, R.E, Kay, L.E. | | Deposit date: | 2002-09-03 | | Release date: | 2002-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Outer Membrane Enzyme PagP by NMR
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7D2O
 
 | |
7V0V
 
 | | GFP Nanobody NMR Structure | | Descriptor: | Anti-GFP Nanobody | | Authors: | Mueller, G.A. | | Deposit date: | 2022-05-11 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nanobody Paratope Ensembles in Solution Characterized by MD Simulations and NMR.
Int J Mol Sci, 23, 2022
|
|
6N8C
 
 | | Structure of the Huntingtin tetramer/dimer mixture determined by paramagnetic NMR | | Descriptor: | Huntingtin | | Authors: | Schwieters, C.D, Kotler, S.A, Schmidt, T, Ceccon, A, Ghirlando, R, Libich, D.S, Clore, G.M. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Probing initial transient oligomerization events facilitating Huntingtin fibril nucleation at atomic resolution by relaxation-based NMR.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8CLR
 
 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | Descriptor: | RNA hairpin with CUUG tetraloop | | Authors: | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
6WA1
 
 | |
8ACZ
 
 | |
1MFJ
 
 | |
