7DVR
 
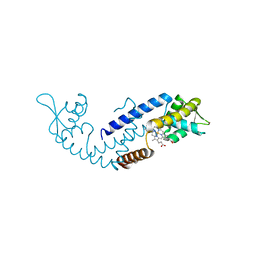 | | Crystal structure of heme sensor protein PefR from Streptococcus agalactiae in complex with heme | | Descriptor: | COBALT (II) ION, HTH marR-type domain-containing protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVV
 
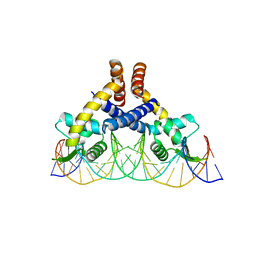 | | Heme sensor protein PefR from Streptococcus agalactiae bound to operator DNA (28-mer) | | Descriptor: | DNA (28-MER), HTH marR-type domain-containing protein | | Authors: | Nishinaga, M, Nagai, S, Nishitani, Y, Sugimoto, H, Shiro, Y, Sawai, H. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
7DVS
 
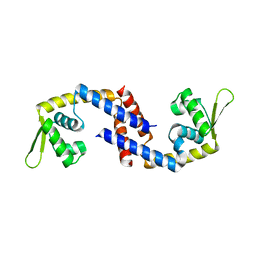 | | Crystal structure of Apo (heme-free) PefR | | Descriptor: | MarR family transcriptional regulator | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
1M43
 
 | |
6KUC
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
6KUD
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 3) | | Descriptor: | GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
6KUB
 
 | | Crystal structure of Plasmodium falciparum histo-aspartic protease (HAP) zymogen (Form 1) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HAP protein | | Authors: | Rathore, I, Mishra, V, Bhaumik, P. | | Deposit date: | 2019-08-31 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation mechanism of plasmepsins, pepsin-like aspartic proteases from Plasmodium, follows a unique trans-activation pathway.
Febs J., 288, 2021
|
|
4EME
 
 | |
4G7L
 
 | | Crystal Structure of rat Heme oxygenase-1 in complex with Heme and O2 | | Descriptor: | FORMIC ACID, Heme oxygenase 1, OXYGEN MOLECULE, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-20 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G99
 
 | | Rat Heme Oxygenase-1 in complex with Heme and CO at 100 K after warming to 160 K | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G8W
 
 | | Rat Heme Oxygenase-1 in complex with Heme and O2 with 13 hr illumination: Laser on | | Descriptor: | FORMIC ACID, Heme oxygenase 1, OXYGEN MOLECULE, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4G98
 
 | | Rat Heme Oxygenase-1 in complex with Heme and CO at 100K | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
4MBA
 
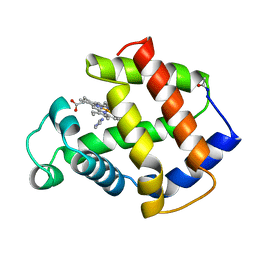 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | IMIDAZOLE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
4M1N
 
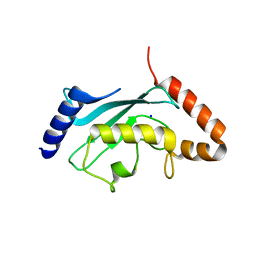 | | Crystal structure of Plasmodium falciparum ubiquitin conjugating enzyme UBC9 | | Descriptor: | SODIUM ION, Ubiquitin conjugating enzyme UBC9 | | Authors: | Boucher, L.E, Reiter, K.H, Bosch, J, Matunis, J.M. | | Deposit date: | 2013-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Biochemically Distinct Properties of the Small Ubiquitin-related Modifier (SUMO) Conjugation Pathway in Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
4OF9
 
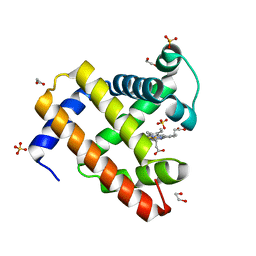 | | Structure of K42N variant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OOD
 
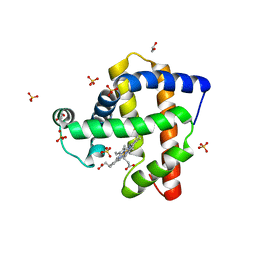 | | Structure of K42Y mutant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8T3U
 
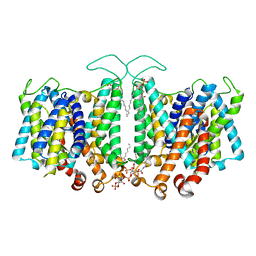 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaCl Buffer: Form2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T47
 
 | |
8T45
 
 | |
8T44
 
 | |
8T3R
 
 | |
8WZM
 
 | |
8WZH
 
 | |
8WZJ
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8WZK
 
 | | Human erythrocyte catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
