7QDQ
 
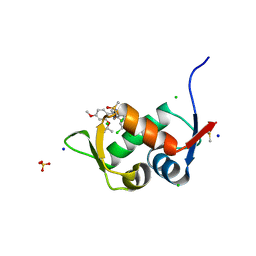 | | Crystal Structure of HDM2 in complex with Caylin-1 | | Descriptor: | CHLORIDE ION, Caylin-1, DIMETHYL SULFOXIDE, ... | | Authors: | Finke, A.D, Walti, M.A, Marsh, M.E, Orts, J. | | Deposit date: | 2021-11-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Elucidation of a nutlin-derivative-HDM2 complex structure at the interaction site by NMR molecular replacement: A straightforward derivation
J Magn Reson Open, 10-11, 2022
|
|
8SS9
 
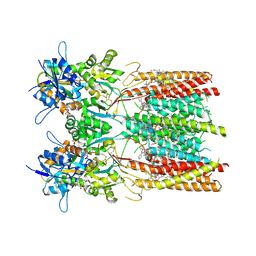 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SXW
 
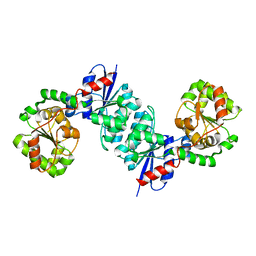 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N mutation, apo structure at pH 6 | | Descriptor: | CHLORIDE ION, SODIUM ION, UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase | | Authors: | Kroft, C.W, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
7QE4
 
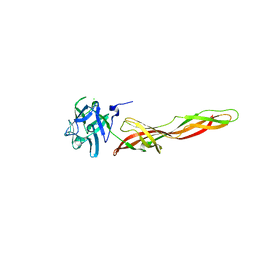 | | B-trefoil lectin from Salpingoeca rosetta in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, ... | | Authors: | Notova, S, Varrot, A. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The choanoflagellate pore-forming lectin SaroL-1 punches holes in cancer cells by targeting the tumor-related glycosphingolipid Gb3.
Commun Biol, 5, 2022
|
|
8SY0
 
 | | X-ray crystal structure of UDP- 2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27 in complex with its product UDP-2,3-diacetamido-2,3-dideoxy-d-mannuronic acid at pH 9 | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | McKnight, J.O, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
7QSF
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-12 (G206C, R207T, D210A, S211A) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase, Isoaspartyl peptidase subunit beta, ... | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-13 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QTC
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-3 (G206H, R207T, D210P, S211Q) | | Descriptor: | Isoaspartyl peptidase, Isoaspartyl peptidase subunit beta, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QVR
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-37 (G206S, R207T, D210S) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QY6
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, wild type (WT EcAIII) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8SYB
 
 | | X-ray crystal structure of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid-2-epimerase from Thermus thermophilus strain HB27, D98N variant in the presence of UDP-2,3-diacetamido-2,3-dideoxy-glucuronic acid and UDP-N-acetylglucosamine at pH 9 | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{R})-4,5-diacetamido-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxane-2-carboxylic acid, CHLORIDE ION, SODIUM ION, ... | | Authors: | Kroft, C.W, Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis.
J.Biol.Chem., 299, 2023
|
|
7TIZ
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-63 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
8SV3
 
 | | 7-Deazapurines and 5-Halogenpyrimidine DNA duplex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Modified DNA, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Conformational Morphing by a DNA Analogue Featuring 7-Deazapurines and 5-Halogenpyrimidines and the Origins of Adenine-Tract Geometry.
Biochemistry, 62, 2023
|
|
7QOA
 
 | | Structure of CodB, a cytosine transporter in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-AMINOPYRIMIDIN-2(1H)-ONE, Cytosine permease, ... | | Authors: | Hatton, C.E, Cameron, A.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of cytosine transport protein CodB provides insight into nucleobase-cation symporter 1 mechanism.
Embo J., 41, 2022
|
|
7QQ8
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-8 (G206Y, R207Q, D210P, S211T) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-01-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYM
 
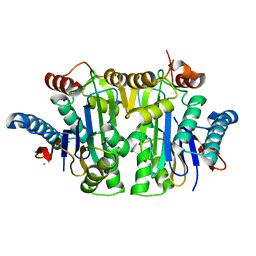 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-18 (R207V, D210P, S211W) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QYX
 
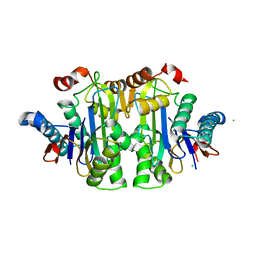 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-24 (R207A, D210S, S211T) | | Descriptor: | Beta-aspartyl-peptidase, CHLORIDE ION, Isoaspartyl peptidase, ... | | Authors: | Loch, J.I, Klonecka, A, Kadziolka, K, Bonarek, P, Barciszewski, J, Imiolczyk, B, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2022-01-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7U34
 
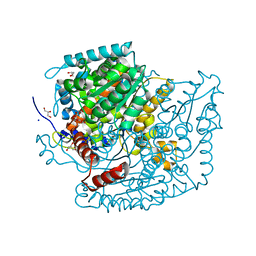 | | The structure of phosphoglucose isomerase from Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Yan, K, Kowalski, B, Fang, W, van Aalten, D. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
7QH0
 
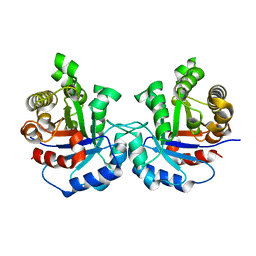 | |
7Q7A
 
 | | Room temperature structure of RNase A at 120 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7QF7
 
 | |
7Q77
 
 | | Room temperature structure of RNase A at 50 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q76
 
 | | Room temperature structure of RNase A at 22 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q79
 
 | | Room temperature structure of RNase A at 100 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q75
 
 | | Room temperature structure of RNase A at atmospheric pressure | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
7Q78
 
 | | Room temperature structure of RNase A at 72 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SODIUM ION, ... | | Authors: | Lieske, J, Guenther, S, Saouane, S, Meents, A. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fixed-target high-pressure macromolecular crystallography
To Be Published
|
|
