4NCZ
 
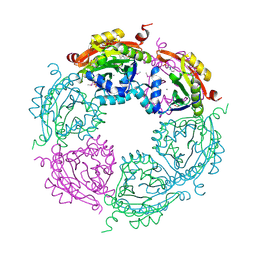 | | Spermidine N-acetyltransferase from Vibrio cholerae in complex with 2-[n-cyclohexylamino]ethane sulfonate. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, SODIUM ION, ... | | Authors: | Osipiuk, J, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-25 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
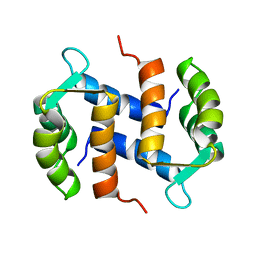
2CNP
 
 | |
2C77
 
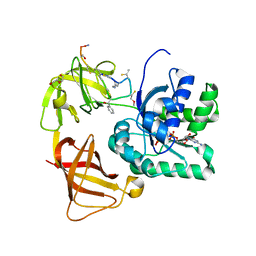 | | EF-Tu complexed with a GTP analog and the antibiotic GE2270 A | | Descriptor: | DI(HYDROXYETHYL)ETHER, ELONGATION FACTOR TU-B, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
4NBN
 
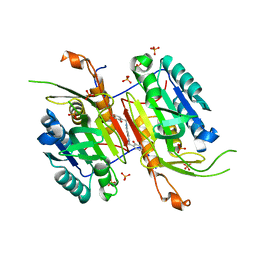 | |
4N5D
 
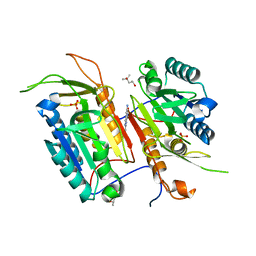 | | Tailoring Small Molecules for an Allosteric Site on Procaspase-6: Cpd1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-amino-2,8-dimethylpyrido[2,3-d]pyrimidin-7(8H)-one, Caspase-6, ... | | Authors: | Murray, J.M, Steffek, M. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tailoring small molecules for an allosteric site on procaspase-6.
Chemmedchem, 9, 2014
|
|
5USN
 
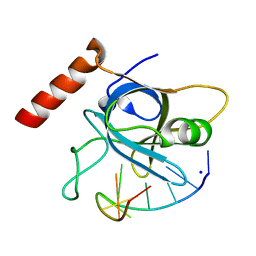 | |
2EVZ
 
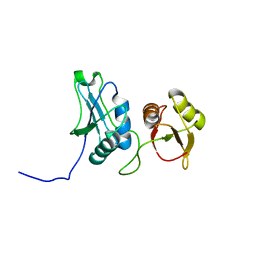 | |
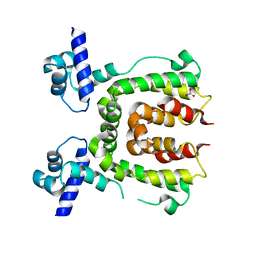
2WV1
 
 | |
3J0A
 
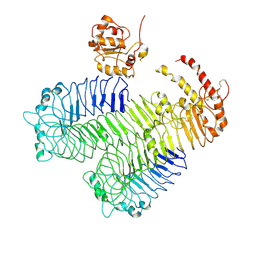 | | Homology model of human Toll-like receptor 5 fitted into an electron microscopy single particle reconstruction | | Descriptor: | Toll-like receptor 5, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Modis, Y, Zhou, K, Kanai, R, Lee, P, Wang, H.W. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin.
J.Struct.Biol., 177, 2012
|
|
4NC4
 
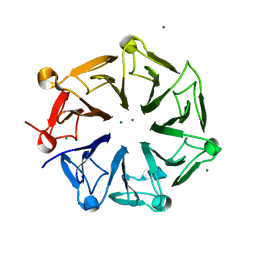 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
3L5X
 
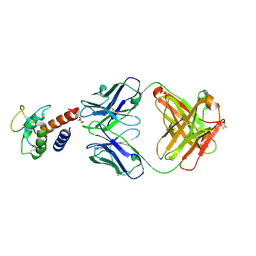 | | Crystal structure of the complex between IL-13 and H2L6 FAB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, H2L6 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
4QTH
 
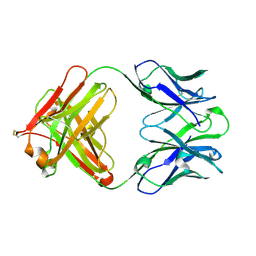 | | Crystal structure of anti-uPAR Fab 8B12 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Zhao, B, Yuan, C, Luo, Z, Huang, M. | | Deposit date: | 2014-07-08 | | Release date: | 2015-02-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation.
J.Mol.Biol., 427, 2015
|
|
2FLO
 
 | |
3RLM
 
 | |
3RSH
 
 | | Structure of 3-ketoacyl-(acyl-carrier-protein)reductase (FabG) from Vibrio cholerae O1 complexed with NADP+ (space group P62) | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hou, J, Chruszcz, M, Cooper, D.R, Grabowski, M, Zheng, H, Osinski, T, Shumilin, I, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-02 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3RMZ
 
 | |
3S3U
 
 | |
2KWD
 
 | |
1R0O
 
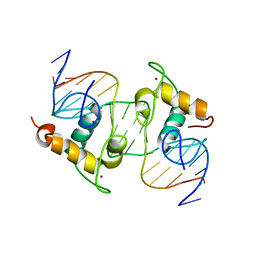 | | Crystal Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex | | Descriptor: | Ecdysone Response Element, Ecdysone receptor, Ultraspiracle protein, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex
Embo J., 22, 2003
|
|
3U09
 
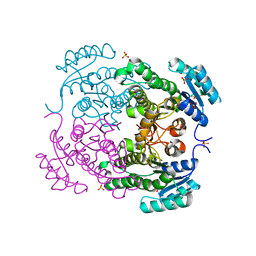 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(G92D) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Fratczak, Z, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3LTM
 
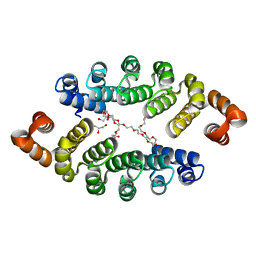 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
3L5W
 
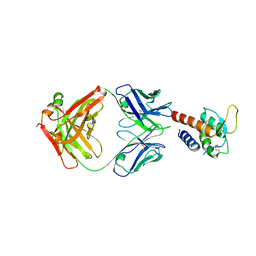 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
3TNF
 
 | | LidA from Legionella in complex with active Rab8a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LidA, MAGNESIUM ION, ... | | Authors: | Schoebel, S, Cichy, A.L, Goody, R.S, Itzen, A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein LidA from Legionella is a Rab GTPase supereffector.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4AD9
 
 | | Crystal structure of human LACTB2. | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE-LIKE PROTEIN 2, ZINC ION | | Authors: | Allerston, C.K, Krojer, T, Shrestha, B, Burgess Brown, N, Chalk, R, Elkins, J.M, Filippakopoulos, P, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Lactb2, a Metallo-Beta-Lactamase Protein, as a Human Mitochondrial Endoribonuclease
Nucleic Acids Res., 44, 2016
|
|
3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
