2H96
 
 | | Discovery of Potent, Highly Selective, and Orally Bioavailable Pyridine Carboxamide C-jun NH2-terminal Kinase Inhibitors | | Descriptor: | 5-CYANO-N-(2,5-DIMETHOXYBENZYL)-6-ETHOXYPYRIDINE-2-CARBOXAMIDE, C-jun-amino-terminal kinase-interacting protein 1, GLYCEROL, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-06-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors.
J.Med.Chem., 49, 2006
|
|
2GMX
 
 | | Selective Aminopyridine-Based C-Jun N-terminal Kinase inhibitors with cellular activity | | Descriptor: | C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, N-(4-AMINO-5-CYANO-6-ETHOXYPYRIDIN-2-YL)-2-(4-BROMO-2,5-DIMETHOXYPHENYL)ACETAMIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-04-07 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aminopyridine-Based c-Jun N-Terminal Kinase Inhibitors with Cellular Activity and Minimal Cross-Kinase Activity.
J.Med.Chem., 49, 2006
|
|
3CX2
 
 | | Crystal structure of the C1 domain of cardiac isoform of myosin binding protein-C at 1.3A | | Descriptor: | Myosin-binding protein C, cardiac-type | | Authors: | Fisher, S.J, Helliwell, J.R, Khurshid, S, Govada, L, Redwood, C, Squire, J.M, Chayen, N.E. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An investigation into the protonation states of the C1 domain of cardiac myosin-binding protein C
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3CP5
 
 | | Cytochrome c from rhodothermus marinus | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Stelter, M, Melo, A, Saraiva, L, Teixeira, M, Archer, M. | | Deposit date: | 2008-03-31 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A novel type of monoheme cytochrome c: biochemical and structural characterization at 1.23 A resolution of rhodothermus marinus cytochrome c
Biochemistry, 47, 2008
|
|
2IN6
 
 | | Wee1 kinase complex with inhibitor PD311839 | | Descriptor: | 3-(9-HYDROXY-1,3-DIOXO-4-PHENYL-2,3-DIHYDROPYRROLO[3,4-C]CARBAZOL-6(1H)-YL)PROPANOIC ACID, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
2IO6
 
 | | Wee1 kinase complexed with inhibitor PD330961 | | Descriptor: | 9-HYDROXY-6-(3-HYDROXYPROPYL)-4-(2-METHOXYPHENYL)PYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
3CX5
 
 | |
7S9Z
 
 | | Helicobacter Hepaticus CcsBA Closed Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
7S9Y
 
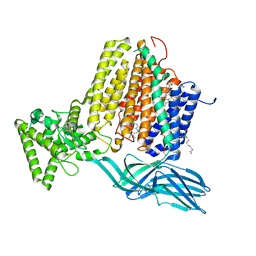 | | Helicobacter Hepaticus CcsBA Open Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
5D98
 
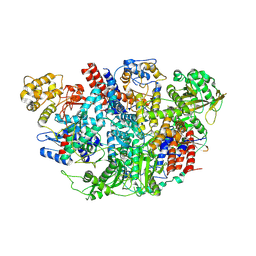 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P43212 | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
2IA8
 
 | | Kinetic and Crystallographic Studies of a Redesigned Manganese-Binding Site in Cytochrome c Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pfister, T, Mirarefi, A.Y, Gengenbach, A.J, Zhao, X, Conaster, C.D.N, Gao, Y.G, Robinson, H, Zukoski, C.F, Wang, A.H.J, Lu, Y. | | Deposit date: | 2006-09-07 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Kinetic and crystallographic studies of a redesigned manganese-binding site in cytochrome c peroxidase
J.Biol.Inorg.Chem., 12, 2007
|
|
5D9A
 
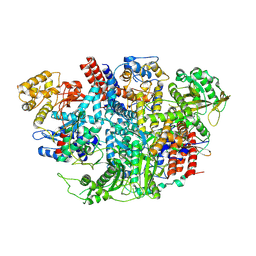 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P212121 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
5Y6G
 
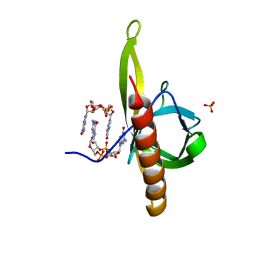 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
2I8F
 
 | |
1LQV
 
 | | Crystal structure of the Endothelial protein C receptor with phospholipid in the groove in complex with Gla domain of protein C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-05-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
3S8G
 
 | | 1.8 A structure of ba3 cytochrome c oxidase mutant (A120F) from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
3S8F
 
 | | 1.8 A structure of ba3 cytochrome c oxidase from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
6YYJ
 
 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P21212 at 2.1 Angstroms | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, C-phycocyanin alpha chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6FU3
 
 | | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae | | Descriptor: | CALCIUM ION, HEME C, Protein CcpR | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae
To Be Published
|
|
8FSJ
 
 | | Cryo-EM structure of engineered hepatitis C virus E1E2 ectodomain in complex with antibodies AR4A, HEPC74, and IGH520 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AR4A heavy chain, ... | | Authors: | Metcalf, M.C, Ofek, G. | | Deposit date: | 2023-01-10 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
Nat Commun, 14, 2023
|
|
8OHM
 
 | |
6YQ8
 
 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup P63 at 1.8 Angstroms | | Descriptor: | 1,2-ETHANEDIOL, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5KE0
 
 | | Discovery of 1-1H-Pyrazolo 4,3-c pyridine-6-yl urea Inhibitors of Extracellular Signal Regulated Kinase ERK for the Treatment of Cancers | | Descriptor: | 1-[3-(2-methylpyridin-4-yl)-1~{H}-pyrazolo[4,3-c]pyridin-6-yl]-3-(phenylmethyl)urea, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Lim, J. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of 1-(1H-Pyrazolo[4,3-c]pyridin-6-yl)urea Inhibitors of Extracellular Signal-Regulated Kinase (ERK) for the Treatment of Cancers.
J.Med.Chem., 59, 2016
|
|
6PFV
 
 | | Structure of S. venezuelae RisG-WhiG-c-di-GMP complex: orthorhombic crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
