6V2D
 
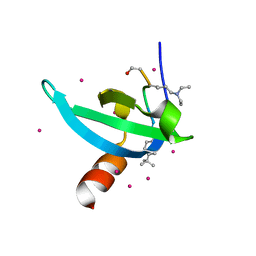 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V41
 
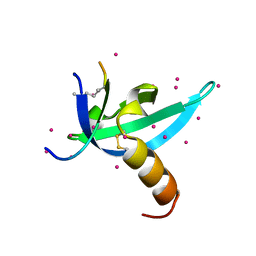 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V8W
 
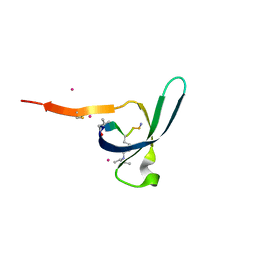 | | CDYL2 chromodomain in complex with a synthetic peptide | | Descriptor: | Chromodomain Y-like protein 2, IVA-PHE-ALA-PHE-5T3-SER-NH2, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, James, L.I, Lamb, K.N, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
2D9U
 
 | | Solution structure of the Chromo domain of chromobox homolog 2 from human | | Descriptor: | Chromobox protein homolog 2 (isoform 2) | | Authors: | Li, H, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chromo domain of chromobox homolog 2 from human
To be Published
|
|
2H1E
 
 | |
2DNT
 
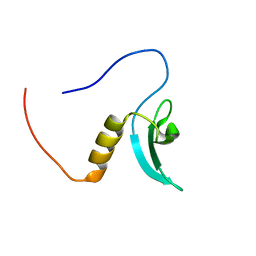 | | Solution Structure of RSGI RUH-064, a Chromo Domain from Human cDNA | | Descriptor: | Chromodomain protein, Y chromosome-like, isoform b | | Authors: | Ruhul Momen, A.Z.M, Hirota, H, Izumi, K, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-064, a Chromo Domain from Human cDNA
to be published
|
|
2DY8
 
 | |
2KVM
 
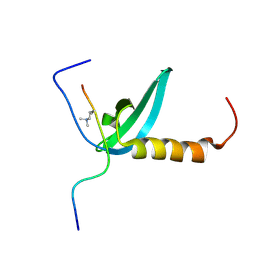 | |
2MJ8
 
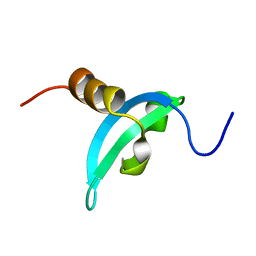 | | Solution structure of CDYL2 chromodomain | | Descriptor: | Chromodomain Y-like protein 2 | | Authors: | Qin, S, Houliston, S, Arrowsmith, C.H, Edwards, A.M, Wu, H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-28 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
2L12
 
 | | Solution NMR structure of the chromobox protein 7 with H3K9me3 | | Descriptor: | Chromobox homolog 7, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Gutmanas, A, Fares, C, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2K28
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 4 | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Fares, C, Gutmanas, A, Ravichandran, M, Loppnau, P, Bountra, C, Weigelt, J, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromo domain of the chromobox protein homolog 4.
To be Published
|
|
2K1B
 
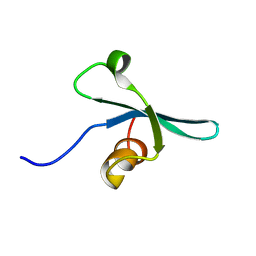 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 7 | | Descriptor: | Chromobox protein homolog 7 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Gutmanas, A, Fares, C, Bountra, C, Weigelt, J, Loppnau, P, Ravichandran, M, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromobox protein homolog 7.
To be Published
|
|
2M2L
 
 | |
2L11
 
 | | Solution NMR structure of the Cbx3 in complex with H3K9me3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2L1B
 
 | | Solution NMR structure of the chromobox protein Cbx7 with H3K27me3 | | Descriptor: | Chromobox protein homolog 7, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Muhandiram, R, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2N88
 
 | | Chromodomain 3 (CD3) of cpSRP43 | | Descriptor: | Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Hennig, J, Sattler, M. | | Deposit date: | 2015-10-06 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
6Q3M
 
 | | Structure of CHD4 PHD2 - tandem chromodomains | | Descriptor: | 1,2-ETHANEDIOL, Chromodomain-helicase-DNA-binding protein 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alt, A, Mancini, E.J. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of histone readers
To Be Published
|
|
6GUU
 
 | |
7NKX
 
 | | RNA polymerase II-Spt4/5-nucleosome-Chd1 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin elongation factor SPT4, ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9EAR
 
 | | CHD1-nucleosome complex (closed state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromodomain-helicase-DNA-binding protein 1, ... | | Authors: | James, A.M, Farnung, L. | | Deposit date: | 2024-11-11 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of human CHD1 nucleosome recruitment and pausing.
Mol.Cell, 85, 2025
|
|
5O9G
 
 | | Structure of nucleosome-Chd1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
9BAZ
 
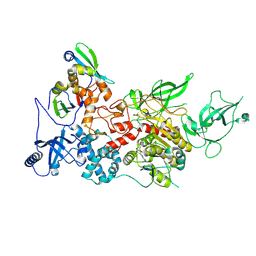 | | CryoEM structure of DIM2-HP1 complex | | Descriptor: | DNA (cytosine-5-)-methyltransferase, Heterochromatin protein one, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9BAQ
 
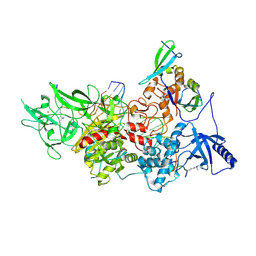 | | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3'), DNA (cytosine-5-)-methyltransferase, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9GD2
 
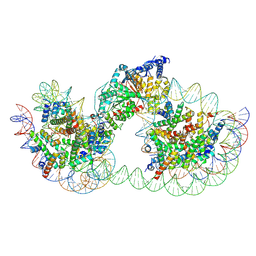 | | Structure of Chd1 bound to a dinucleosome with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
9GD1
 
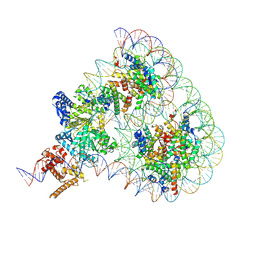 | | Structure of Chd1 bound to a hexasome-nucleosome complex with a dyad-to-dyad distance of 103 bp. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Engeholm, M, Roske, J.J, Oberbeckmann, E, Dienemann, C, Lidschreiber, M, Cramer, P, Farnung, L. | | Deposit date: | 2024-08-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resolution of transcription-induced hexasome-nucleosome complexes by Chd1 and FACT.
Mol.Cell, 84, 2024
|
|
