1IX5
 
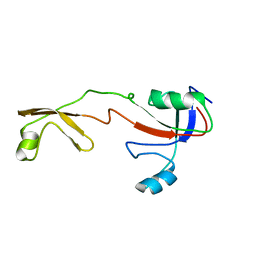 | | Solution structure of the Methanococcus thermolithotrophicus FKBP | | Descriptor: | FKBP | | Authors: | Suzuki, R, Nagata, K, Kawakami, M, Nemoto, N, Furutani, M, Adachi, K, Maruyama, T, Tanokura, M. | | Deposit date: | 2002-06-12 | | Release date: | 2003-06-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional Solution Structure of an Archaeal FKBP with a Dual Function of Peptidyl Prolyl cis-trans Isomerase and Chaperone-like Activities
J.MOL.BIOL., 328, 2003
|
|
1J4H
 
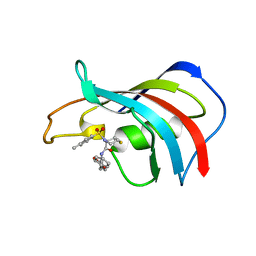 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1D6O
 
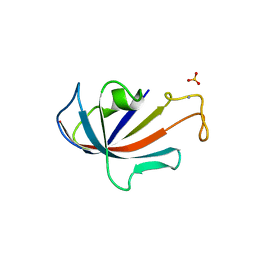 | | NATIVE FKBP | | Descriptor: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-15 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
7F2J
 
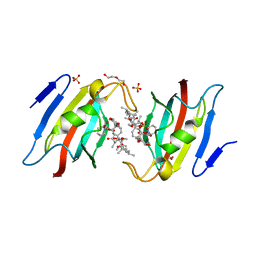 | | Crystal structure of AtFKBP53 FKBD in complex with rapamycin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase FKBP53, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Singh, A.K, Saharan, K, Vasudevan, D. | | Deposit date: | 2021-06-11 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal packing reveals rapamycin-mediated homodimerization of an FK506-binding domain.
Int.J.Biol.Macromol., 206, 2022
|
|
4ODK
 
 | |
4ODR
 
 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, ACETATE ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
1D7H
 
 | | FKBP COMPLEXED WITH DMSO | | Descriptor: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7J
 
 | | FKBP COMPLEXED WITH 4-HYDROXY-2-BUTANONE | | Descriptor: | 4-HYDROXY-2-BUTANONE, AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
7DKI
 
 | | Silk worm FKBP, isoform-1 | | Descriptor: | GLYCEROL, Peptidylprolyl isomerase | | Authors: | Yuchi, Z, Nayak, B.C. | | Deposit date: | 2020-11-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Silk worm FKBP, isoform-1
To Be Published
|
|
1D7I
 
 | | FKBP COMPLEXED WITH METHYL METHYLSULFINYLMETHYL SULFIDE (DSS) | | Descriptor: | AMMONIUM ION, METHYL METHYLSULFINYLMETHYL SULFIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1J4I
 
 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1L1P
 
 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
7ETT
 
 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit QFPFV | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
7ETV
 
 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit DFPFV | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
7ETU
 
 | | The FK1 domain of FKBP51 in complex with peptide-inhibitor hit SFPFT | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, peptide-inhibitor hit | | Authors: | Han, J.T, Zhu, Y.C, Pan, D.B, Xue, H.X, Wang, S, Liu, H.X, He, Y.X, Yao, X.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of pentapeptide-inhibitor hits targeting FKBP51 by combining computational modeling and X-ray crystallography.
Comput Struct Biotechnol J, 19, 2021
|
|
1JVW
 
 | | TRYPANOSOMA CRUZI MACROPHAGE INFECTIVITY POTENTIATOR (TCMIP) | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Pereira, P.J.B, Vega, M.C, Gonzalez-Rey, E, Fernandez-Carazo, R, Macedo-Ribeiro, S, Gomis-Rueth, F.X, Gonzalez, A, Coll, M. | | Deposit date: | 2001-08-31 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Trypanosoma cruzi macrophage infectivity potentiator has a rotamase core and a highly exposed alpha-helix.
EMBO Rep., 3, 2002
|
|
4TW6
 
 | | The Fk1 domain of FKBP51 in complex with iFit1 | | Descriptor: | (3-{(1R)-3-(3,4-dimethoxyphenyl)-1-[({(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)pent-4-enoyl]piperidin-2-yl}carbonyl)oxy]propyl}phenoxy)acetic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
4TW8
 
 | | The Fk1-Fk2 domains of FKBP52 in complex with iFit-FL | | Descriptor: | 2-(5-{[({3-[(1R)-1-[({(2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetyl)amino]methyl}-6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
1N1A
 
 | | Crystal Structure of the N-terminal domain of human FKBP52 | | Descriptor: | FKBP52 | | Authors: | Li, P, Ding, Y, Wu, B, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-terminal domain of human FKBP52.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2LPV
 
 | |
2MF9
 
 | | Solution structure of the N-terminal domain of human FKBP38 (FKBP38NTD) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP8 | | Authors: | Kang, C, Ye, H, Simon, B, Sattler, M, Yoon, H.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Functional role of the flexible N-terminal extension of FKBP38 in catalysis.
Sci Rep, 3, 2013
|
|
4MGV
 
 | |
2M2A
 
 | |
4LAV
 
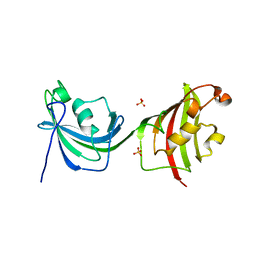 | | Crystal Structure Analysis of FKBP52, Crystal Form II | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP4, SULFATE ION | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
4LAX
 
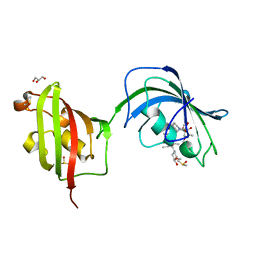 | | Crystal Structure Analysis of FKBP52, Complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Bracher, A, Kozany, C, Haehle, A, Wild, P, Zacharias, M, Hausch, F. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Crystal Structures of the Free and Ligand-Bound FK1-FK2 Domain Segment of FKBP52 Reveal a Flexible Inter-Domain Hinge.
J.Mol.Biol., 425, 2013
|
|
