8VMZ
 
 | |
8VN2
 
 | |
8WFV
 
 | |
9BCZ
 
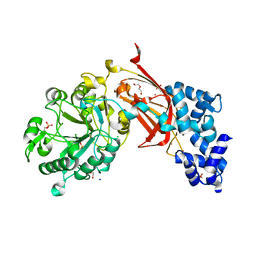 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
8VN7
 
 | |
8VNA
 
 | |
8Z76
 
 | | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2 during 6 months | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, SODIUM ION, ... | | Authors: | Varfolomeeva, L.A, Solovieva, A.Y, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-04-19 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of thiocyanate dehydrogenase from Pelomicrobium methylotrophicum (pmTcDH), activated by crystals soaking with 1 mM CuCl2
To Be Published
|
|
8VMV
 
 | |
8VNL
 
 | |
8VNQ
 
 | |
8VMY
 
 | |
8QTG
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 9) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-5-(trifluoromethyl)-1~{H}-pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
8YOU
 
 | | The pmTcDH complex structure with an inhibitor SeCN | | Descriptor: | COPPER (II) ION, GLYCEROL, SELENIUM ATOM, ... | | Authors: | Varfolomeeva, L.A, Polyakov, K.M, Shipkov, N.S, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2024-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The pmTcDH complex structure with an inhibitor SeCN
To Be Published
|
|
8VN3
 
 | |
8QTH
 
 | | Crystal structure of CBL-b in complex with an allosteric inhibitor (compound 8) | | Descriptor: | 1-methyl-5-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-(trifluoromethyl)-7H-pyrrolo[2,3-b]pyridin-6-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
9EQH
 
 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the Inhibitor Space of the WWP1 and WWP2 HECT E3 Ligases
To Be Published
|
|
8VNF
 
 | |
8VNG
 
 | |
8VUI
 
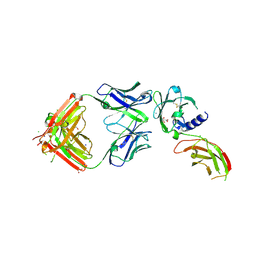 | | Structure of FabS1CE-EPR-1, an elbow-locked Fab, in complex with the erythropoeitin receptor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-29 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
9B1Z
 
 | |
8VNH
 
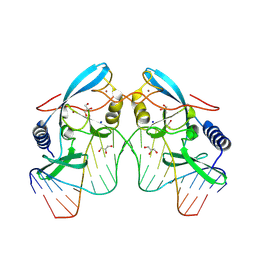 | |
8VNN
 
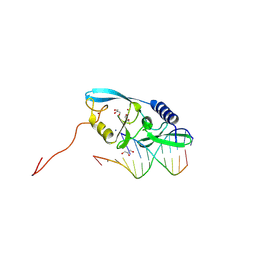 | |
8PTE
 
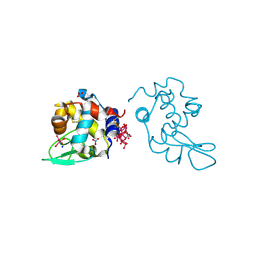 | | Polyoxidovanadate interaction with proteins: crystal structure of lysozyme bound to octadecavanadate ion (structure B) | | Descriptor: | Lysozyme C, NITRATE ION, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Ferraro, G, Merlino, A. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Stabilization and Binding of [V 4 O 12 ] 4- and Unprecedented [V 20 O 54 (NO 3 )] n- to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug V IV O(acetylacetonato) 2.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8VNR
 
 | |
8QTJ
 
 | | Crystal structure of Cbl-b in complex with an allosteric inhibitor (compound 30) | | Descriptor: | 3-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-1-[(1~{R})-1-(1-methylpyrazol-4-yl)ethyl]-5-(trifluoromethyl)pyridin-2-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2023-10-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Discovery, Optimization, and Biological Evaluation of Arylpyridones as Cbl-b Inhibitors.
J.Med.Chem., 67, 2024
|
|
