7NE2
 
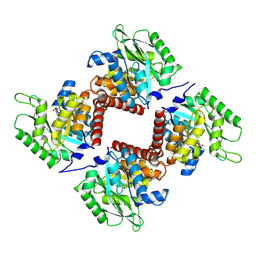 | | Crystal structure of class I SFP aldolase YihT from Salmonella enterica with SFP/ DHAP (Schiff base complex with active site Lys193) | | Descriptor: | (2~{S},3~{S},4~{R})-2,3,4,5-tetrakis(oxidanyl)-6-phosphonooxy-hexane-1-sulfonic acid, Sulfofructosephosphate aldolase, [(~{E})-2,3-bis(oxidanyl)prop-1-enyl] dihydrogen phosphate | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
6G8G
 
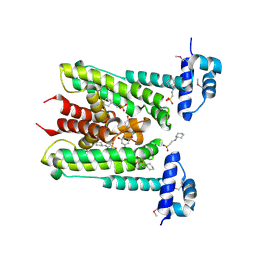 | | Flavonoid-responsive Regulator FrrA in complex with Genistein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GENISTEIN, TetR/AcrR family transcriptional regulator | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
7TEC
 
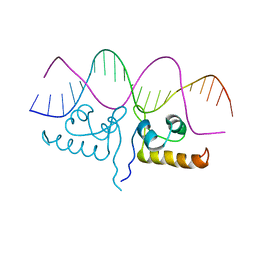 | |
3LSJ
 
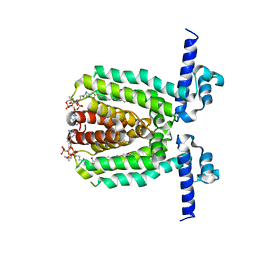 | |
3LSR
 
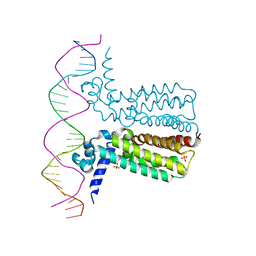 | |
3LSP
 
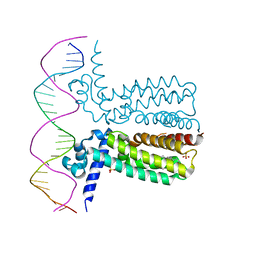 | | Crystal Structure of DesT bound to desCB promoter and oleoyl-CoA | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*TP*CP*GP*AP*GP*TP*CP*AP*AP*CP*AP*AP*GP*CP*GP*TP*TP*CP*AP*CP*TP*GP*AP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*AP*TP*CP*AP*GP*TP*GP*AP*AP*CP*GP*CP*TP*TP*GP*TP*TP*GP*AP*CP*TP*CP*GP*AP*TP*TP*G)-3'), DesT, ... | | Authors: | Miller, D.J, White, S.W. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for the transcriptional regulation of membrane lipid homeostasis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6G8H
 
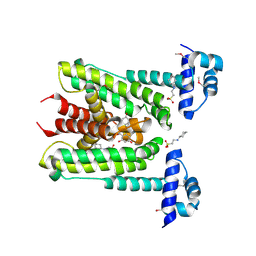 | | Flavonoid-responsive Regulator FrrA in complex with (R,S)-Naringenin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, NARINGENIN, R-naringenin, ... | | Authors: | Werner, N, Hoppen, J, Palm, G, Werten, S, Goettfert, M, Hinrichs, W. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The induction mechanism of the flavonoid-responsive regulator FrrA.
Febs J., 2021
|
|
3MWH
 
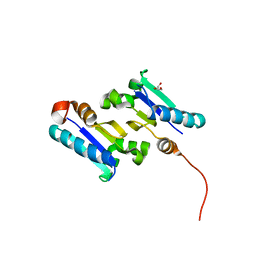 | | The 1.4 Ang crystal structure of the ArsD arsenic metallochaperone provides insights into its interactions with the ArsA ATPase | | Descriptor: | Arsenical resistance operon trans-acting repressor arsD, GLYCEROL | | Authors: | Ye, J, Ajees, A.A, Yang, J, Rosen, B.P. | | Deposit date: | 2010-05-05 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The 1.4 A crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase.
Biochemistry, 49, 2010
|
|
4DO2
 
 | |
6R1P
 
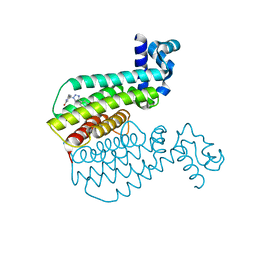 | | EthR ligand complex | | Descriptor: | 2-[2-[4-(2,3-dihydro-1,4-benzodioxin-6-yl)-1,2,3-triazol-1-yl]ethyl]-6-methyl-1~{H}-pyrimidin-4-one, HTH-type transcriptional regulator EthR | | Authors: | Pohl, E, Tatum, N. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relative Binding Energies Predict Crystallographic Binding Modes of Ethionamide Booster Lead Compounds.
J Phys Chem Lett, 10, 2019
|
|
6R1S
 
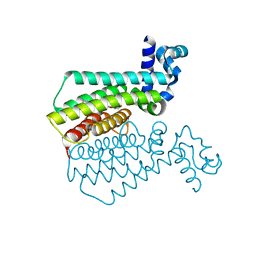 | | EthR ligand complex | | Descriptor: | 2-(3-methylphenyl)-~{N}-[[2-(oxan-4-yl)-7-oxidanyl-pyrazolo[1,5-a]pyrimidin-5-yl]methyl]ethanamide, HTH-type transcriptional regulator EthR | | Authors: | Pohl, E, Tatum, N. | | Deposit date: | 2019-03-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relative Binding Energies Predict Crystallographic Binding Modes of Ethionamide Booster Lead Compounds.
J Phys Chem Lett, 10, 2019
|
|
4RRU
 
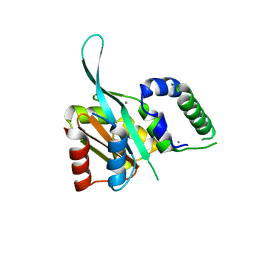 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
6EK9
 
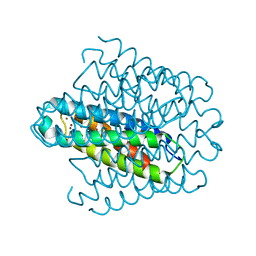 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
7ZG6
 
 | | TacA1 antitoxin | | Descriptor: | DUF1778 domain-containing protein, MAGNESIUM ION | | Authors: | Grabe, G.J, Morgan, R.M.L, Helaine, S. | | Deposit date: | 2022-04-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular stripping underpins derepression of a toxin-antitoxin system.
Nat.Struct.Mol.Biol., 2024
|
|
7ZG5
 
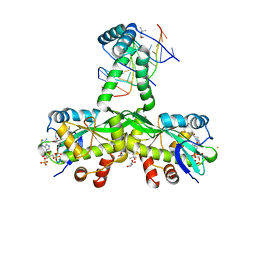 | | The crystal structure of Salmonella TacAT3-DNA complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetyltransferase, BARIUM ION, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Helaine, S. | | Deposit date: | 2022-04-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular stripping underpins derepression of a toxin-antitoxin system.
Nat.Struct.Mol.Biol., 2024
|
|
5NZ2
 
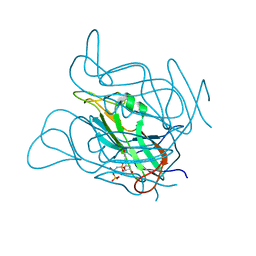 | | Twist and induce: Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 dUTPase. D95E mutant from dUTPase 80alpha phage. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Alite, C, Humphrey, S, Donderis, J, Maiques, E, Ciges-Tomas, J.R, Penades, J.R, Marina, A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 alpha dUTPase.
Sci Rep, 7, 2017
|
|
5NYZ
 
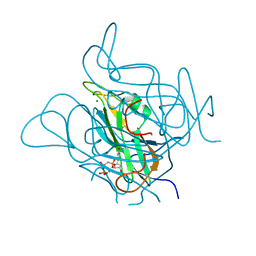 | | Twist and induce: Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 dUTPase. D95E mutant from dUTPase 80alpha phage. | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPase, MAGNESIUM ION | | Authors: | Alite, C, Humphrey, S, Donderis, J, Maiques, E, Ciges-Tomas, J.R, Penades, J.R, Marina, A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissecting the link between the enzymatic activity and the SaPI inducing capacity of the phage 80 alpha dUTPase.
Sci Rep, 7, 2017
|
|
7R5A
 
 | |
3Q76
 
 | | Structure of human neutrophil elastase (uncomplexed) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Neutrophil elastase, ... | | Authors: | Hansen, G, Niefind, K. | | Deposit date: | 2011-01-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Unexpected active-site flexibility in the structure of human neutrophil elastase in complex with a new dihydropyrimidone inhibitor.
J.Mol.Biol., 409, 2011
|
|
3Q77
 
 | | Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl (4R)-4-(4-cyanophenyl)-6-methyl-2-oxo-1-[3-(trifluoromethyl)phenyl]-1,2,3,4-tetrahydropyrimidine-5-carboxylate, ... | | Authors: | Hansen, G, Niefind, K. | | Deposit date: | 2011-01-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Unexpected active-site flexibility in the structure of human neutrophil elastase in complex with a new dihydropyrimidone inhibitor.
J.Mol.Biol., 409, 2011
|
|
8G24
 
 | |
8G25
 
 | |
8G26
 
 | |
6CDA
 
 | | Crystal structure of L34A CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, GLYCEROL, ... | | Authors: | Capdevila, D.A, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
4RS9
 
 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-07 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
