6L9T
 
 | | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Viswanathan, V, Pandey, N, Singh, A, Sinha, M, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with OSCN at 1.89 A resolution
To Be Published
|
|
6B37
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,3-benzothiazol-2-ol | | Descriptor: | 1,3-benzothiazol-2-ol, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,3-benzothiazol-2-ol
To Be Published
|
|
4XZM
 
 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
6BHR
 
 | | HIV-1 immature CTD-SP1 hexamer in complex with IP6 | | Descriptor: | Capsid protein p24,Spacer peptide 1, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6E5Y
 
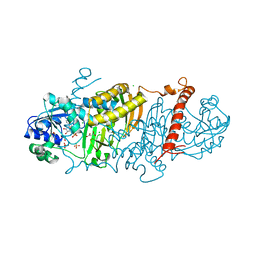 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
4ZN6
 
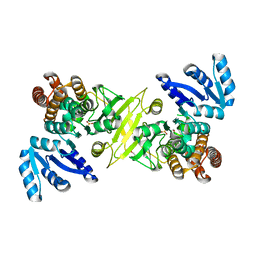 | |
4WYY
 
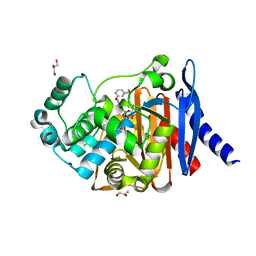 | | Crystal Structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
6MNC
 
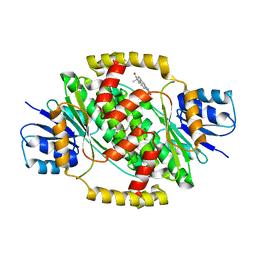 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, DI(HYDROXYETHYL)ETHER, Estradiol 17-beta-dehydrogenase 1 | | Authors: | Li, T, Stephen, P, Zhu, D.W, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
7EZP
 
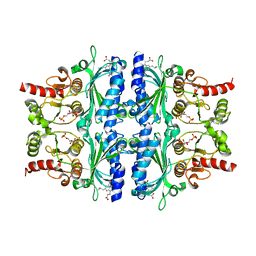 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
6MNE
 
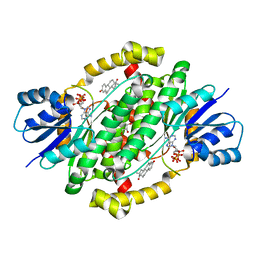 | | CRYSTAL STRUCTURE OF HUMAN 17BETA-HYDROXYSTEROID DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRONE AND NADP+ | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estradiol 17-beta-dehydrogenase 1, GLYCEROL, ... | | Authors: | Li, T, Lin, S.X. | | Deposit date: | 2018-10-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of human 17 beta-hydroxysteroid dehydrogenase type 1 complexed with estrone and NADP+reveal the mechanism of substrate inhibition.
Febs J., 286, 2019
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
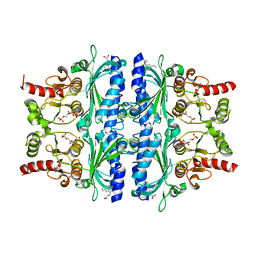 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
2YBB
 
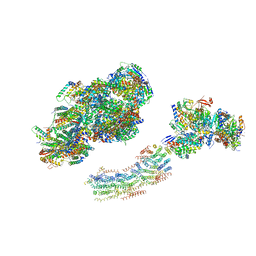 | | Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Althoff, T, Mills, D.J, Popot, J.-L, Kuehlbrandt, W. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Embo J., 30, 2011
|
|
6KDM
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:entecavir 5'-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6KDN
 
 | | HIV-1 reverse transcriptase with Q151M/Y115F/F116Y:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2019-07-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural features in common of HBV and HIV-1 resistance against chirally-distinct nucleoside analogues entecavir and lamivudine.
Sci Rep, 10, 2020
|
|
6FSD
 
 | | Mus musculus acetylcholinesterase in complex with 2-(4-Biphenylyloxy)-N-[3-(1-piperidinyl)propyl]-acetamide hydrochloride (10) | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(4-phenylphenoxy)-~{N}-(3-piperidin-1-ylpropyl)ethanamide, ... | | Authors: | Knutsson, S, Engdahl, C, Kumari, R, Kindahl, T, Forsgren, N, Lindgren, C, Kitur, S, Kamau, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Inhibitors of Mosquito Acetylcholinesterase 1 with Resistance-Breaking Potency.
J.Med.Chem., 61, 2018
|
|
3KK6
 
 | | Crystal Structure of Cyclooxygenase-1 in complex with celecoxib | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[5-(4-METHYLPHENYL)-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL]BENZENESULFONAMIDE, CITRATE ANION, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2009-11-04 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Coxibs interfere with the action of aspirin by binding tightly to one monomer of cyclooxygenase-1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5DPX
 
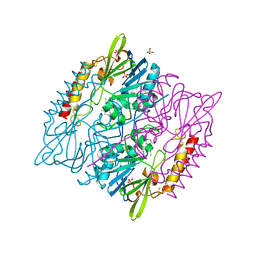 | | 1,2,4-Triazole-3-thione compounds as inhibitors of L1, di-zinc metallo-beta-lactamases. | | Descriptor: | 5-(2-methylphenyl)-3H-1,2,4-triazole-3-thione, Metallo-beta-lactamase L1 type 3, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Khan, R, Dideberg, O. | | Deposit date: | 2015-09-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases.
ChemMedChem, 12, 2017
|
|
6MCS
 
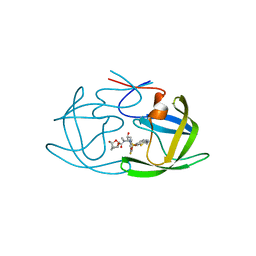 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
4P2F
 
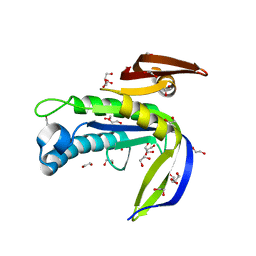 | | Monomeric form of a single mutant (F363R) of Mycobacterial Adenylyl cyclase Rv1625c | | Descriptor: | 1,2-ETHANEDIOL, Adenylate cyclase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Barathy, D.V, Mattoo, R, Visweswariah, S.S, Suguna, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | New structural forms of a mycobacterial adenylyl cyclase Rv1625c.
Iucrj, 1, 2014
|
|
3HHY
 
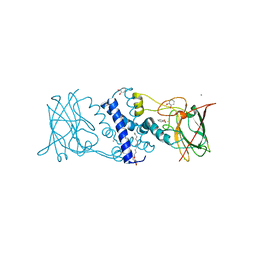 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with catechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CATECHOL, CHLORIDE ION, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
5Z5Q
 
 | | Nukacin ISK-1 in active state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
3HJ8
 
 | | Crystal structure determination of catechol 1,2-dioxygenase from rhodococcus opacus 1CP in complex with 4-chlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 4-CHLOROBENZENE-1,2-DIOL, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-21 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts
J.Struct.Biol., 170, 2010
|
|
3HJQ
 
 | | Crystal structure of catechol 1,2-dioxygenase from Rhodococcus opacus 1CP in complex with 3-methylcatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3-METHYLCATECHOL, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3HJS
 
 | | Crystal structure of catechol 1,2-dioxygenase from Rhodococcus opacus 1CP in complex with 4-methylcatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 4-METHYLCATECHOL, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
