7W3E
 
 | | Bovine cytochrome c oxidese in CN-bound fully reduced state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
2N8A
 
 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
7VS7
 
 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
7VZV
 
 | | Active state CI from Q10 dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W00
 
 | | Deactive state CI from Q10 dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-12-14 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0R
 
 | | Active state CI from Q10-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W1O
 
 | | Deactive state CI from Q10-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-12-21 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W2K
 
 | | Deactive state CI from Rotenone-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-01-25 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W1V
 
 | | Active state CI from Rotenone-NADH dataset, Subclass 1 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-20 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4J
 
 | | Deactive state CI from Q1-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W1T
 
 | | Active state CI from Rotenone dataset, Subclass 1 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-20 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W31
 
 | | Deactive state CI from DQ-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4C
 
 | | Active state CI from Q1-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-27 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W2R
 
 | | Active state CI from DQ-NADH dataset, Subclass 1 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6P2I
 
 | | Acyclic imino acid reductase (Bsp5) in complex with NADPH and D-Arg | | Descriptor: | 1,2-ETHANEDIOL, D-ARGININE, Glycerate dehydrogenase, ... | | Authors: | Guo, J, Higgins, M.A, Daniel-Ivad, P, Ryan, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | An Asymmetric Reductase That Intercepts Acyclic Imino Acids Producedin Situby a Partner Oxidase.
J.Am.Chem.Soc., 141, 2019
|
|
7PH8
 
 | |
4XPC
 
 | |
4XPE
 
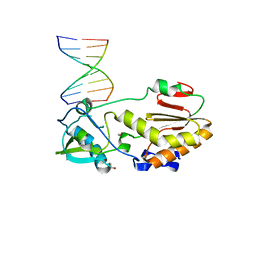 | |
7OVZ
 
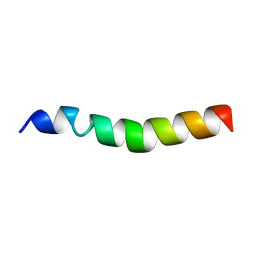 | |
7LVA
 
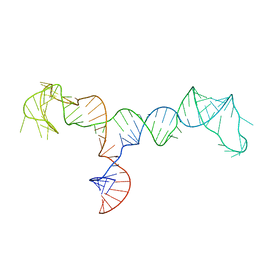 | | Solution structure of the HIV-1 PBS-segment | | Descriptor: | RNA (103-MER) | | Authors: | Heng, X, Song, Z. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | The three-way junction structure of the HIV-1 PBS-segment binds host enzyme important for viral infectivity.
Nucleic Acids Res., 49, 2021
|
|
4UTG
 
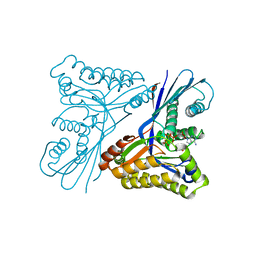 | | Burkholderia pseudomallei heptokinase WcbL,AMPPNP (ATP analogue) complex. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
8ORK
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | De Rose, S.A, Isupov, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
4X1T
 
 | | The crystal structure of Arabidopsis thaliana galactolipid synthase MGD1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Monogalactosyldiacylglycerol synthase 1, chloroplastic, ... | | Authors: | Rocha, J, Breton, C. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants.
Plant J., 85, 2016
|
|
4X3J
 
 | | Selection of fragments for kinase inhibitor design: decoration is key | | Descriptor: | 1-[4-(4-amino-5-oxopyrido[2,3-d]pyrimidin-8(5H)-yl)phenyl]-3-[2-fluoro-5-(trifluoromethyl)phenyl]urea, Angiopoietin-1 receptor | | Authors: | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | Deposit date: | 2014-11-30 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
7Q2D
 
 | | mycolic acid methyltransferase Hma (MmaA4) from Mycobac-terium tuberculosis in complex with ZT320 | | Descriptor: | 1,2-ETHANEDIOL, 2-METHOXY-5-METHYLANILINE, DIMETHYL SULFOXIDE, ... | | Authors: | Maveyraud, L, Galy, R, Mourey, L. | | Deposit date: | 2021-10-25 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fragment-Based Ligand Discovery Applied to the Mycolic Acid Methyltransferase Hma (MmaA4) from Mycobacterium tuberculosis : A Crystallographic and Molecular Modelling Study.
Pharmaceuticals, 14, 2021
|
|
