3A9E
 
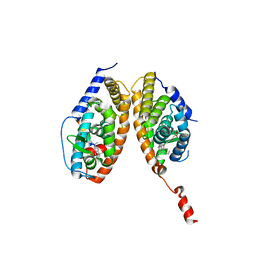 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | Authors: | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-10-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
8VJB
 
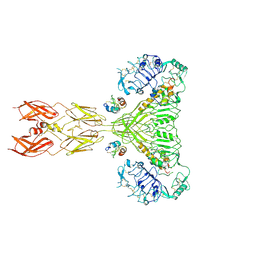 | | Cryo-EM structure of short form insulin receptor (IR-A) with four IGF2 bound, symmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
7P4N
 
 | |
8VJC
 
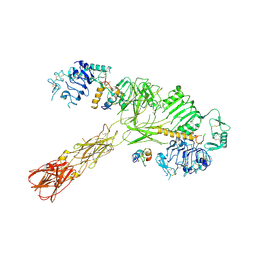 | | Cryo-EM structure of short form insulin receptor (IR-A) with three IGF2 bound, asymmetric conformation. | | Descriptor: | Insulin-like growth factor II, Isoform Short of Insulin receptor | | Authors: | An, W, Hall, C, Li, J, Huang, A, Wu, J, Park, J, Bai, X.C, Choi, E. | | Deposit date: | 2024-01-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation of the insulin receptor by insulin-like growth factor 2.
Nat Commun, 15, 2024
|
|
9BZ7
 
 | |
384D
 
 | |
8FQP
 
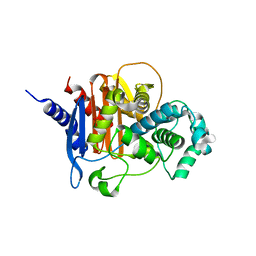 | | apo ADC-162 beta-lactamase | | Descriptor: | Beta-lactamase, GLYCINE, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fernando, M.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
2XGS
 
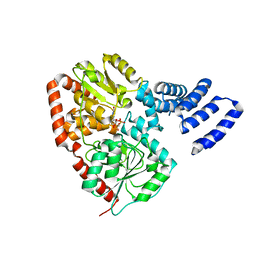 | | XcOGT in complex with C-UDP | | Descriptor: | 5'-O-[(S)-HYDROXY(PHOSPHONOMETHYL)PHOSPHORYL]URIDINE, XCOGT | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Blair, D.E, Pathak, S, Navratilova, I, van Aalten, D.M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Substrate and Product Analogues as Human O-Glcnac Transferase Inhibitors.
Amino Acids, 40, 2011
|
|
8DFO
 
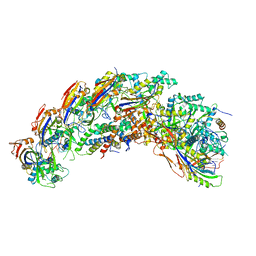 | | type I-C Cascade bound to AcrIC4 | | Descriptor: | AcrIC4, CRISPR-associated protein, CT1133 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
1EVP
 
 | |
8FQQ
 
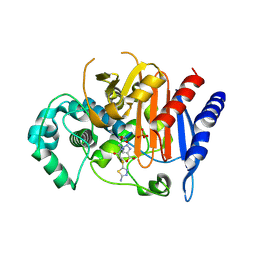 | | ADC-162 in complex with boronic acid transition state inhibitor MB076 | | Descriptor: | 1-[(2R)-2-{2-[(5-amino-1,3,4-thiadiazol-2-yl)sulfanyl]acetamido}-2-boronoethyl]-1H-1,2,3-triazole-4-carboxylic acid, Beta-lactamase, GLYCINE, ... | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fernando, M.C. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
8DEJ
 
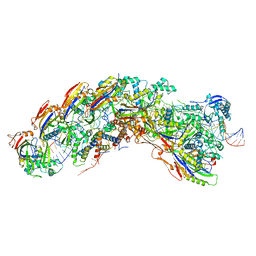 | | D. vulgaris type I-C Cascade bound to dsDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-20 | | Release date: | 2023-02-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
6TYT
 
 | |
6J8N
 
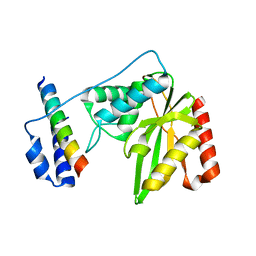 | | Crystal structure of SVBP-VASH1 complex, mutation C169A of VASH1 | | Descriptor: | Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Liao, S, Gao, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis.
Cell Res., 29, 2019
|
|
6J9H
 
 | |
7C4D
 
 | | Marine microorganism esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative esterase, ... | | Authors: | Zhu, C.H, Wu, Y.K, Isupov, M.N. | | Deposit date: | 2020-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into a Novel Esterase from the East Pacific Rise and Its Improved Thermostability by a Semirational Design.
J.Agric.Food Chem., 69, 2021
|
|
2AJ4
 
 | | Crystal structure of Saccharomyces cerevisiae Galactokinase in complex with galactose and Mg:AMPPNP | | Descriptor: | CHLORIDE ION, Galactokinase, MAGNESIUM ION, ... | | Authors: | Thoden, J.B, Sellick, C.A, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer
J.Biol.Chem., 280, 2005
|
|
6RKW
 
 | | CryoEM structure of the complete E. coli DNA Gyrase complex bound to a 130 bp DNA duplex | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (58-MER), DNA (62-MER), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
5KVF
 
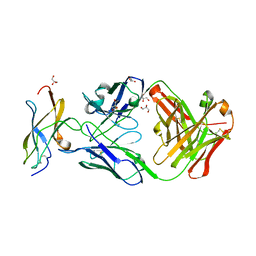 | | Zika specific antibody, ZV-64, bound to ZIKA envelope DIII | | Descriptor: | GLYCEROL, ZV-64 Antibody Fab Heavy Chain, ZV-64 Antibody Fab Light Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
5OGA
 
 | | Structure of minimal i-motif domain | | Descriptor: | DNA (5'-D(*TP*(DCP)P*GP*TP*TP*CP*(DCP)P*GP*TP*TP*TP*TP*TP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Mir, B, Serrano, I, Buitrago, D, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2017-07-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH.
J. Am. Chem. Soc., 139, 2017
|
|
2WFH
 
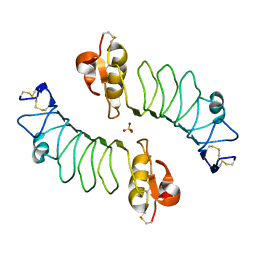 | | The Human Slit 2 Dimerization Domain D4 | | Descriptor: | SLIT HOMOLOG 2 PROTEIN C-PRODUCT, SULFATE ION | | Authors: | Seiradake, E, von Philipsborn, A.C, Henry, M, Fritz, M, Lortat-Jacob, H, Jamin, M, Hemrika, W, Bastmeyer, M, Cusack, S, McCarthy, A.A. | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and functional relevance of the Slit2 homodimerization domain.
EMBO Rep., 10, 2009
|
|
4XEI
 
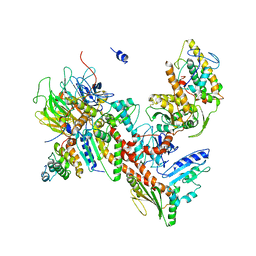 | | Orthorhombic isomorph of bovine Arp2/3 complex | | Descriptor: | Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, Actin-related protein 2/3 complex subunit 2, ... | | Authors: | Jurgenson, C.J, Pollard, T.P. | | Deposit date: | 2014-12-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.874 Å) | | Cite: | Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4ZQV
 
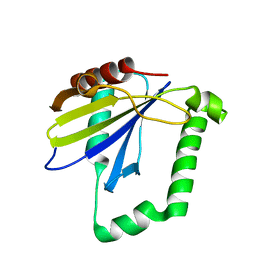 | |
7SXK
 
 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - Full Length | | Descriptor: | Portal protein | | Authors: | Hou, C.F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
6UGC
 
 | | C3 symmetric peptide design number 3 | | Descriptor: | C3-3 cyclic peptide design, CADMIUM ION, SODIUM ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-26 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
