8INJ
 
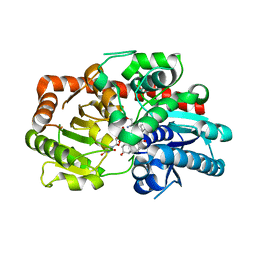 | | Crystal structure of UGT74AN3-UDP-DIG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | Authors: | Feng, L, Wei, H. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of UGT74AN3-UDP-DIG
To Be Published
|
|
6VN6
 
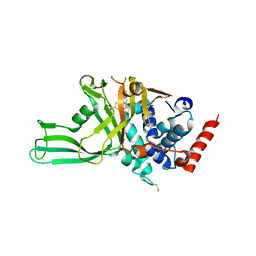 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 14 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION, [(2R)-5-chloro-7-{2-[(2S)-1-chloro-2,3-dihydroxypropan-2-yl]thieno[3,2-b]pyridin-7-yl}-2,3-dihydro-1-benzofuran-2-yl](piperazin-1-yl)methanone | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
2GTM
 
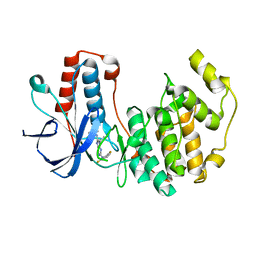 | | Mutated Mouse P38 MAP Kinase Domain in complex with Inhibitor PG-892579 | | Descriptor: | 8-(2-CHLOROPHENYLAMINO)-2-(2,6-DIFLUOROPHENYLAMINO)-9-ETHYL-9H-PURINE-1,7-DIIUM, Mitogen-activated protein kinase 14 | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Sabat, M. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The development of novel C-2, C-8, and N-9 trisubstituted purines as inhibitors of TNF-alpha production.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7EDD
 
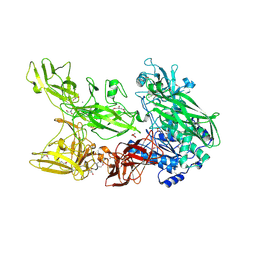 | |
6G5W
 
 | | Crystal Structure of KDM4A with compound YP-03-038 | | Descriptor: | (4~{R})-5-methyl-4-phenyl-2-pyridin-2-yl-pyrazolidin-3-one, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | Malecki, P.H, Carter, D.M, Gohlke, U, Specker, E, Nazare, M, Weiss, M.S, Heinemann, U. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Enhanced Properties of a Benzimidazole Benzylpyrazole Lysine Demethylase Inhibitor: Mechanism-of-Action, Binding Site Analysis, and Activity in Cellular Models of Prostate Cancer.
J.Med.Chem., 64, 2021
|
|
7BC4
 
 | | Cryo-EM structure of fatty acid synthase (FAS) from Pichia pastoris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Snowden, J.S, Alzahrani, J, Sherry, L, Stacey, M, Rowlands, D.J, Ranson, N.A, Stonehouse, N.J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Pichia pastoris fatty acid synthase.
Sci Rep, 11, 2021
|
|
6FD5
 
 | | Crystal Structure of Human APRT-Tyr105Phe variant in complex with Adenine, PRPP and Mg2+, 14 days post crystallization (with AMP and PPi products partially generated) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, ... | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
6U3K
 
 | | The crystal structure of 4-(pyridin-2-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-2-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
7BPI
 
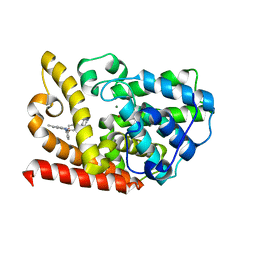 | | The crystal structue of PDE10A complexed with 14 | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2020-03-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4000864 Å) | | Cite: | Discovery of highly selective and orally available benzimidazole-based phosphodiesterase 10 inhibitors with improved solubility and pharmacokinetic properties for treatment of pulmonary arterial hypertension.
Acta Pharm Sin B, 10, 2020
|
|
6MYN
 
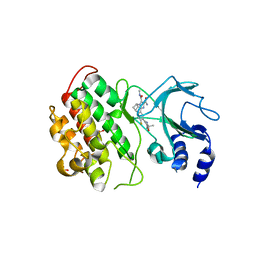 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to inhibitor R7 | | Descriptor: | (5s,7s)-9-fluoro-10-[(3R)-3-hydroxy-3-(5-methyl-1,2-oxazol-3-yl)but-1-yn-1-yl]-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Harris, S.F, Smith, M, Barker, J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Structure Based Design of Potent Selective Inhibitors of Protein Kinase D1 (PKD1).
Acs Med.Chem.Lett., 10, 2019
|
|
8K79
 
 | |
6FD4
 
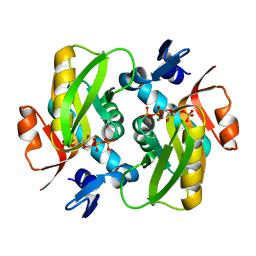 | | Crystal Structure of Human APRT-Tyr105Phe variant in complex with Adenine, PRPP and Mg2+, 14 hours post crystallization | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Adenine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
8K78
 
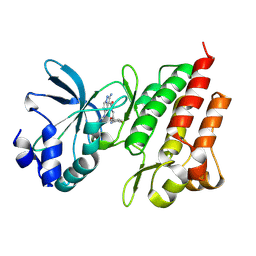 | |
6N2Z
 
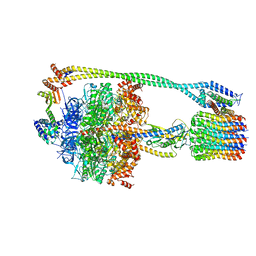 | | Bacillus PS3 ATP synthase class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
2HP8
 
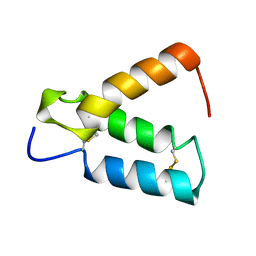 | | SOLUTION STRUCTURE OF HUMAN P8-MTCP1, A CYSTEINE-RICH PROTEIN ENCODED BY THE MTCP1 ONCOGENE,REVEALS A NEW ALPHA-HELICAL ASSEMBLY MOTIF, NMR, 30 STRUCTURES | | Descriptor: | Cx9C motif-containing protein 4 | | Authors: | Barthe, P, Chiche, L, Strub, M.P, Roumestand, C. | | Deposit date: | 1997-08-26 | | Release date: | 1998-03-04 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human p8MTCP1, a cysteine-rich protein encoded by the MTCP1 oncogene, reveals a new alpha-helical assembly motif.
J.Mol.Biol., 274, 1997
|
|
6N30
 
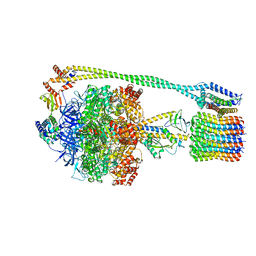 | | Bacillus PS3 ATP synthase class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6VSE
 
 | |
2GPS
 
 | | Crystal Structure of the Biotin Carboxylase Subunit, E23R mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
2GTN
 
 | | Mutated MAP kinase P38 (Mus Musculus) in complex with Inhbitor PG-951717 | | Descriptor: | 2-(2,6-DIFLUOROPHENOXY)-N-(2-FLUOROPHENYL)-9-ISOPROPYL-9H-PURIN-8-AMINE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Sabat, M. | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The development of novel C-2, C-8, and N-9 trisubstituted purines as inhibitors of TNF-alpha production.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6FXX
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Gal, Hg2+ Soak | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6UWR
 
 | |
6N1K
 
 | |
6UWT
 
 | |
7NIN
 
 | | X-ray crystal structure of LsAA9A - CinnamtanninB1 soak | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Auxiliary activity 9, ... | | Authors: | Frandsen, K.E.H, Tokin, R, Skov, L, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of lytic polysaccharide monooxygenase by natural plant extracts.
New Phytol., 232, 2021
|
|
7NIM
 
 | | X-ray crystal structure of LsAA9A - cinnamon extract soak | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Auxiliary activity 9, ... | | Authors: | Frandsen, K.E.H, Tokin, R, Skov, L, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of lytic polysaccharide monooxygenase by natural plant extracts.
New Phytol., 232, 2021
|
|
