6QZ5
 
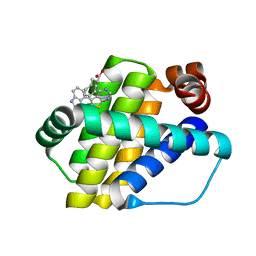 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
8QM8
 
 | | Potential drug binding sites for translation initiation factor eIF4E | | Descriptor: | (2~{R})-2-[(1~{S})-1-[4-(2-fluorophenyl)-2-(2-hydroxyethylamino)phenyl]propoxy]propan-1-ol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Cleasby, A. | | Deposit date: | 2023-09-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Integrating fragment-based screening with targeted protein degradation and genetic rescue to explore eIF4E function.
Nat Commun, 15, 2024
|
|
8QM7
 
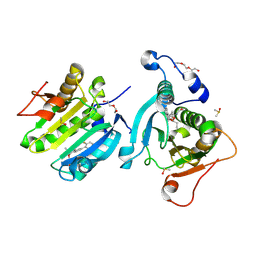 | | Potential drug binding sites for translation initiation factor eIF4E | | Descriptor: | (2~{R})-2-[(1~{R})-1-[4-(2-fluorophenyl)-2-(2-hydroxyethylamino)phenyl]propoxy]propan-1-ol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Cleasby, A. | | Deposit date: | 2023-09-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Integrating fragment-based screening with targeted protein degradation and genetic rescue to explore eIF4E function.
Nat Commun, 15, 2024
|
|
4GG1
 
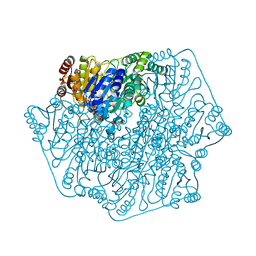 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403T | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-04 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
8QM5
 
 | | Potential drug binding sites for translation initiation factor eIF4E | | Descriptor: | 1-(4-chlorophenyl)cyclopentane-1-carboxylic acid, Eukaryotic translation initiation factor 4E, TRIETHYLENE GLYCOL | | Authors: | Cleasby, A. | | Deposit date: | 2023-09-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Integrating fragment-based screening with targeted protein degradation and genetic rescue to explore eIF4E function.
Nat Commun, 15, 2024
|
|
8QM9
 
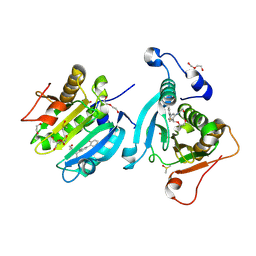 | | Potential drug binding sites for translation initiation factor eIF4E | | Descriptor: | (2~{R})-2-[(1~{S})-1-[4-(2-fluorophenyl)-2-(2-hydroxyethylamino)phenyl]propoxy]propan-1-ol, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cleasby, A. | | Deposit date: | 2023-09-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Integrating fragment-based screening with targeted protein degradation and genetic rescue to explore eIF4E function.
Nat Commun, 15, 2024
|
|
8QM6
 
 | | Potential drug binding sites for translation initiation factor eIF4E | | Descriptor: | (1~{R})-1-[4-(2-fluorophenyl)phenyl]ethanol, (1~{S})-1-[4-(2-fluorophenyl)phenyl]ethanol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cleasby, A. | | Deposit date: | 2023-09-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Integrating fragment-based screening with targeted protein degradation and genetic rescue to explore eIF4E function.
Nat Commun, 15, 2024
|
|
8QM4
 
 | | Potential drug binding sites for translation initiation factor eIF4E | | Descriptor: | 3-phenylphenol, DIMETHYL SULFOXIDE, Eukaryotic translation initiation factor 4E, ... | | Authors: | Cleasby, A. | | Deposit date: | 2023-09-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Integrating fragment-based screening with targeted protein degradation and genetic rescue to explore eIF4E function.
Nat Commun, 15, 2024
|
|
4KCJ
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with N,N'-((ethane-1,2-diylbis(oxy))bis(3,1-phenylene))bis(thiophene-2-carboximidamide) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N,N'-[ethane-1,2-diylbis(oxybenzene-3,1-diyl)]dithiophene-2-carboximidamide, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
4KFF
 
 | | Crystal structure of Hansenula polymorpha copper amine oxidase-1 reduced by methylamine at pH 8.5 | | Descriptor: | COPPER (II) ION, GLYCEROL, Peroxisomal primary amine oxidase | | Authors: | Johnson, B.J, Yukl, E.T, Klema, V.J, Wilmot, C.M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural evidence for the semiquinone in a copper amine oxidase from Hansenula polymorpha: implications for the catalytic mechanism
J.Biol.Chem., 2013
|
|
4KGL
 
 | | Crystal structure of human alpha-L-iduronidase complex with [2R,3R,4R,5S]-2-carboxy-3,4,5-trihydroxy-piperidine | | Descriptor: | (2R,3R,4R,5S)-3,4,5-trihydroxypiperidine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bie, H, Yin, J, He, X, Kermode, A.R, Goddard-Borger, E.D, Withers, S.G, James, M.N.G. | | Deposit date: | 2013-04-29 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Insights into mucopolysaccharidosis I from the structure and action of alpha-L-iduronidase.
Nat.Chem.Biol., 9, 2013
|
|
6QQ5
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric oxide reductase subunit B, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
4FVZ
 
 | | Structure of rat nNOS heme domain in complex with N(omega)-methyl- N(omega)-methoxy-L-arginine | | Descriptor: | (5E)-5-[(N-methoxy-N-methylcarbamimidoyl)imino]-L-norvaline, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Methylated N(omega)-Hydroxy-l-arginine Analogues as Mechanistic Probes for the Second Step of the Nitric Oxide Synthase-Catalyzed Reaction
Biochemistry, 52, 2013
|
|
5IYD
 
 | | Human core-PIC in the initial transcribing state (no IIS) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
4K5B
 
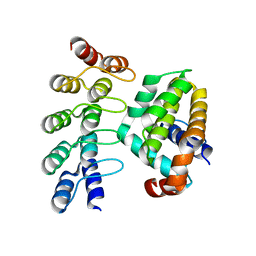 | | Co-crystallization with conformation-specific designed ankyrin repeat proteins explains the conformational flexibility of BCL-W | | Descriptor: | Apoptosis regulator BCL-W, Bcl-2-like protein 2 | | Authors: | Schilling, J, Schoeppe, J, Sauer, E, Plueckthun, A. | | Deposit date: | 2013-04-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-Crystallization with Conformation-Specific Designed Ankyrin Repeat Proteins Explains the Conformational Flexibility of BCL-W
J.Mol.Biol., 426, 2014
|
|
2X1S
 
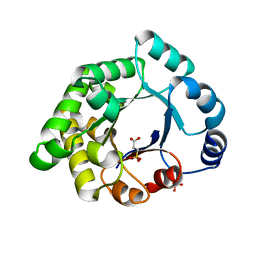 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 3-SULFOPROPANOIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Castejeijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6QYO
 
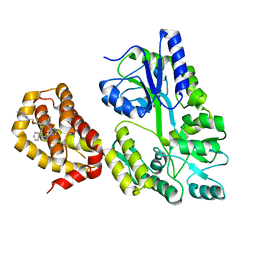 | | Structure of MBP-Mcl-1 in complex with compound 18a | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
2X1U
 
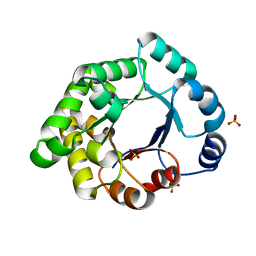 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4KCI
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with N,N'-(ethane-1,2-diylbis(3,1-phenylene))bis(thiophene-2-carboximidamide) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N,N'-(ethane-1,2-diyldibenzene-3,1-diyl)dithiophene-2-carboximidamide, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
5J5Y
 
 | | Translation initiation factor 4E in complex with m2(7,2'O)GppCCl2ppG mRNA 5' cap analog | | Descriptor: | 2-amino-9-{5-O-[(R)-{[(S)-{dichloro[(R)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-2-O-methyl-beta-D-ribofuranosyl}-7-methyl-9H-purin-7-ium-6-olate, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-04 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
4F5K
 
 | |
5ZIE
 
 | |
6STJ
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Cystatin domain-containing protein, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
5ZJZ
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
4F5G
 
 | |
