7P0V
 
 | | Crystal structure of human SF3A1 ubiquitin-like domain in complex with U1 snRNA stem-loop 4 | | Descriptor: | Isoform 2 of Splicing factor 3A subunit 1, PENTAETHYLENE GLYCOL, RNA (5'-R(P*GP*GP*GP*GP*AP*CP*UP*GP*CP*GP*UP*UP*CP*GP*CP*GP*CP*UP*UP*UP*CP*CP*CP*C)-3'), ... | | Authors: | Sabath, K, de Vries, T, Jonas, S. | | Deposit date: | 2021-06-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Sequence-specific RNA recognition by an RGG motif connects U1 and U2 snRNP for spliceosome assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7P6F
 
 | | 1.93 A resolution X-ray crystal structure of the transcriptional regulator SrnR from Streptomyces griseus | | Descriptor: | ACETATE ION, SODIUM ION, Transcriptional regulator SrnR | | Authors: | Mazzei, L, Ciurli, S. | | Deposit date: | 2021-07-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure, dynamics, and function of SrnR, a transcription factor for nickel-dependent gene expression.
Metallomics, 13, 2021
|
|
7OL7
 
 | |
7OL6
 
 | |
7P8P
 
 | | Crystal structure of Fhit covalently bound to a nucleotide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bis(5'-adenosyl)-triphosphatase, SODIUM ION, ... | | Authors: | Herzog, D, Missun, M, Diederichs, K, Marx, A. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Chemical Proteomics of the Tumor Suppressor Fhit Covalently Bound to the Cofactor Ap 3 A Elucidates Its Inhibitory Action on Translation.
J.Am.Chem.Soc., 144, 2022
|
|
7OPH
 
 | | NaK S-DI mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-31 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OOK
 
 | | Bacteriophage PRD1 Major Capsid Protein P3 in complex with CPZ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CHLORIDE ION, ... | | Authors: | Duyvesteyn, H.M.E, Peccati, F, Martinez-Castillo, A, Jimenez-Oses, G, Oksanen, H.M, Stuart, D.I, Abrescia, N.G.A. | | Deposit date: | 2021-05-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Bacteriophage PRD1 as a nanoscaffold for drug loading
Nanoscale, 13, 2021
|
|
7OPT
 
 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | Ascorbate peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
6O8G
 
 | | Crystal structure of UvrB bound to fully duplex DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*CP*GP*AP*TP*GP*GP*AP*GP*A)-3'), ... | | Authors: | Lee, S.-J, Sung, R.-J, Verdine, G.L. | | Deposit date: | 2019-03-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Mechanism of DNA Lesion Homing and Recognition by the Uvr Nucleotide Excision Repair System.
Res, 2019, 2019
|
|
7OQR
 
 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | ACETATE ION, Ascorbate peroxidase, GLYCEROL, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
7P26
 
 | |
7OXX
 
 | | CrabP2 mutant R30AK31A | | Descriptor: | Cellular retinoic acid-binding protein 2, SODIUM ION | | Authors: | Tomlinson, C.W.E, Basle, A, Pohl, E. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural requirements for the specific binding of CRABP2 to cyclin D3
To Be Published
|
|
7P23
 
 | |
6ON9
 
 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, tetragonal form | | Descriptor: | NeuRonal IgCAM-5, SODIUM ION, SULFATE ION, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OA8
 
 | | Superfolder Green Fluorescent Protein with 4-cyano-L-phenylalanine at the chromophore (position 66) | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, SODIUM ION, ... | | Authors: | Piacentini, J, Olenginski, G.M, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and spectrophotometric investigation of two unnatural amino-acid altered chromophores in the superfolder green fluorescent protein
Acta Crystallogr.,Sect.D, 2021
|
|
7P0J
 
 | | Crystal structure of S.pombe Mdb1 BRCT domains | | Descriptor: | CITRIC ACID, DNA damage response protein Mdb1, MAGNESIUM ION, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Phosphorylation-dependent assembly of DNA damage response systems and the central roles of TOPBP1.
DNA Repair (Amst), 108, 2021
|
|
7PCO
 
 | | BurG E232Q mutant (holo) in complex with 2R,3R-2,3-dihydroxy-6-methyl-heptanoate (12): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (2R,3R)-6-methyl-2,3-bis(oxidanyl)heptanoic acid, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
7PCM
 
 | | BurG (holo) in complex with (Z)-2,3-dihydroxy-6-methyl-hept-2-enoate (13): Biosynthesis of cyclopropanol rings in bacterial toxins | | Descriptor: | (Z)-6-methyl-2,3-bis(oxidanyl)hept-2-enoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, ... | | Authors: | Trottmann, F, Ishida, K, Ishida, M, Kries, H, Groll, M, Hertweck, C. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead.
Nat.Chem., 14, 2022
|
|
6OMG
 
 | | Structure of mouse CD1D- Glc-DAG (sn-1 C18:0, sn-2 C18:1c9)-iNKT TCR Ternary complex | | Descriptor: | (2R)-1-(alpha-D-glucopyranosyloxy)-3-(octadecanoyloxy)propan-2-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirk, M.Z, Bitra, A, Wang, J. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of mouse CD1D- Glc-DAG (sn-1 C18:0, sn-2 C18:1c9)-iNKT TCR Ternary complex
To Be Published
|
|
7PHX
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - acid-stable sulfotyrosine analogue | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Pereira, P.J.B, Ripoll-Rozada, J, Calisto, B.M. | | Deposit date: | 2021-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and evaluation of peptidic thrombin inhibitors bearing acid-stable sulfotyrosine analogues.
Chem.Commun.(Camb.), 57, 2021
|
|
7Q47
 
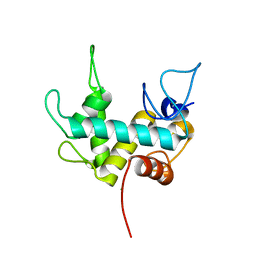 | | Endolysin from bacteriophage Enc34, catalytic domain | | Descriptor: | CHLORIDE ION, Endolysin, SODIUM ION | | Authors: | Cernooka, E, Rumnieks, J, Kazaks, A, Tars, K. | | Deposit date: | 2021-10-29 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diversity of the lysozyme fold: structure of the catalytic domain from an unusual endolysin encoded by phage Enc34.
Sci Rep, 12, 2022
|
|
7PF8
 
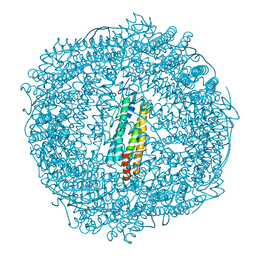 | | SynFtn Variant E141A | | Descriptor: | CHLORIDE ION, Ferritin, SODIUM ION | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Key carboxylate residues for iron transit through the prokaryotic ferritin Syn Ftn.
Microbiology (Reading, Engl.), 167, 2021
|
|
7PFJ
 
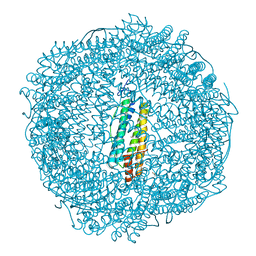 | |
7PFI
 
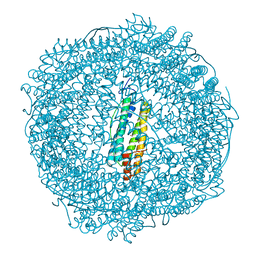 | |
7PRP
 
 | |
