4DQN
 
 | |
2JDW
 
 | |
3TAT
 
 | | TYROSINE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, TYROSINE AMINOTRANSFERASE | | Authors: | Ko, T.P, Yang, W.Z, Wu, S.P, Tsai, H, Yuan, H.S. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of the Escherichia coli tyrosine aminotransferase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4NOG
 
 | | Crystal structure of a putative ornithine aminotransferase from Toxoplasma gondii ME49 in complex with pyrodoxal-5'-phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Halavaty, A, Ruan, J, Shuvalova, L, Flores, K, Dubrovska, I, Ngo, H, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
2HGW
 
 | |
3PDX
 
 | | Crystal structural of mouse tyrosine aminotransferase | | Descriptor: | Tyrosine aminotransferase | | Authors: | Mehere, P.V, Han, Q, Lemkul, J.A, Robinson, H, Bevan, D.R, Li, J. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
Protein Cell, 1, 2010
|
|
4MA8
 
 | | Crystal structure of mouse prion protein complexed with Chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Major prion protein, POM1 heavy chain, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2013-08-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of prion inhibition by phenothiazine compounds.
Structure, 22, 2014
|
|
6J2V
 
 | | GABA aminotransferase from Corynebacterium glutamicum | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]BUTANOIC ACID, GLYCEROL, PLP-dependent aminotransferases | | Authors: | Hong, J, Kim, K.J. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase in complex with a PLP-GABA adduct from Corynebacterium glutamicum.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5X7H
 
 | | Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose | | Descriptor: | CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, MALONATE ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Suzuki, R, Momma, M, Funane, K. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isomaltooligosaccharide-binding structure ofPaenibacillussp. 598K cycloisomaltooligosaccharide glucanotransferase
Biosci. Rep., 37, 2017
|
|
4XBM
 
 | | X-ray crystal structure of Notch ligand Delta-like 1 | | Descriptor: | Delta-like protein 1, alpha-L-fucopyranose | | Authors: | Kershaw, N.J, Burgess, A.W, Church, N.L, Luo, C.S, Adam, T.E. | | Deposit date: | 2014-12-17 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Notch ligand delta-like1: X-ray crystal structure and binding affinity.
Biochem.J., 468, 2015
|
|
4DBC
 
 | | Substrate Activation in Aspartate Aminotransferase | | Descriptor: | (E)-N-{2-hydroxy-3-methyl-6-[(phosphonooxy)methyl]benzylidene}-L-aspartic acid, 1,2-ETHANEDIOL, Aspartate aminotransferase, ... | | Authors: | Toney, M.D, Fisher, A.J, Griswold, W.R. | | Deposit date: | 2012-01-14 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ground-state electronic destabilization via hyperconjugation in aspartate aminotransferase.
J.Am.Chem.Soc., 134, 2012
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | Descriptor: | GLYCEROL, Putative aminotransferase | | Authors: | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
7MFO
 
 | | X-ray structure of the L136 Aminotransferase from Acanthamoeba polyphaga mimivirus in the presence of TDP and PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFP
 
 | | crystal structure of the L136 aminotransferase K185A from acanthamoeba polyphaga mimivirus in the presence of the UDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-dihydroxy-5-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-methyloxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
7MFQ
 
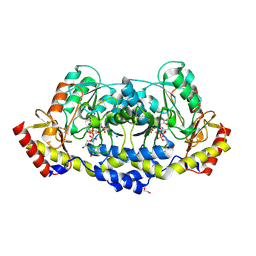 | | crystal structure of the L136 aminotransferase from acanthamoeba polyphaga mimivirus in complex with the TDP-viosamine external aldimine | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Seltzner, C.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2021-04-10 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of an aminotransferase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 30, 2021
|
|
2JJG
 
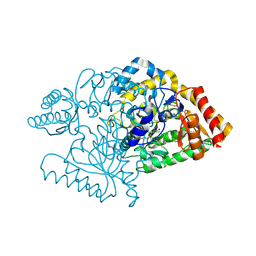 | |
5YB0
 
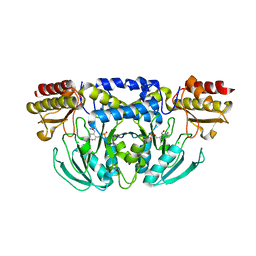 | |
1YAA
 
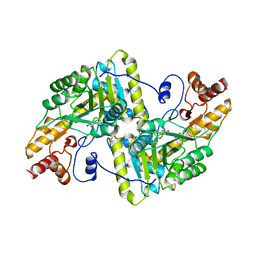 | |
5G08
 
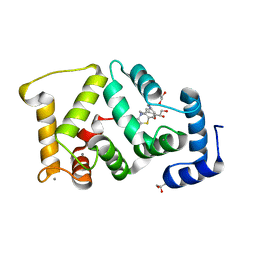 | | Crystal structure of Drosophila NCS-1 bound to chlorpromazine | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MEE
 
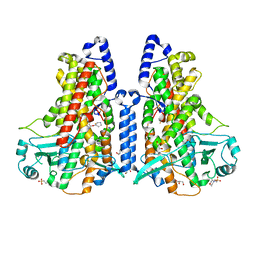 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304V | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Arachidonate 15-lipoxygenase, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
5WPH
 
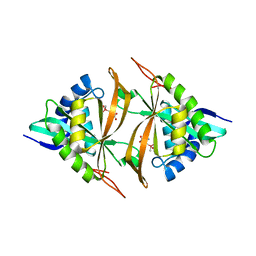 | | Crystal structure of ArsN, N-acetyltransferase with substrate AST from Pseudomonas putida KT2440 | | Descriptor: | (2S)-2-amino-4-[hydroxy(methyl)arsoryl]butanoic acid, Phosphinothricin N-acetyltransferase, SODIUM ION | | Authors: | Venkadesh, S, Dheeman, D.S, Yoshinaga, M, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2017-08-04 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
5YD2
 
 | |
4NEV
 
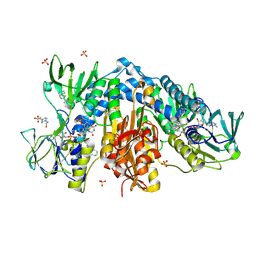 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
5MEF
 
 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304F | | Descriptor: | Arachidonate 15-lipoxygenase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
5YII
 
 | |
