5VMY
 
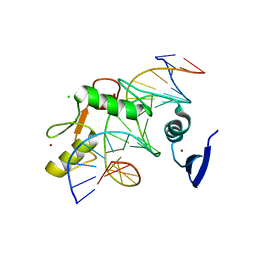 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a hemi CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*CP*GP*CP*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
9F3D
 
 | |
8FD4
 
 | |
4UAM
 
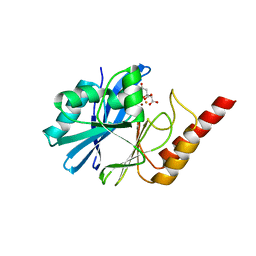 | | 1.8 Angstrom crystal structure of IMP-1 metallo-beta-lactamase with a mixed iron-zinc center in the active site | | Descriptor: | CITRATE ANION, FE (III) ION, IMP-1 metallo-beta-lactamase, ... | | Authors: | Carruthers, T.J, Carr, P.D, Jackson, C.J, Otting, G. | | Deposit date: | 2014-08-11 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Iron(III) Located in the Dinuclear Metallo-beta-Lactamase IMP-1 by Pseudocontact Shifts.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6DR3
 
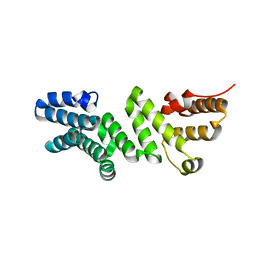 | | Crystal structure of E. coli LpoA amino terminal domain | | Descriptor: | Penicillin-binding protein activator LpoA | | Authors: | Kelley, A.C, Saper, M.A. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structures of the amino-terminal domain of LpoA from Escherichia coli and Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5L85
 
 | |
5L1C
 
 | |
7QUI
 
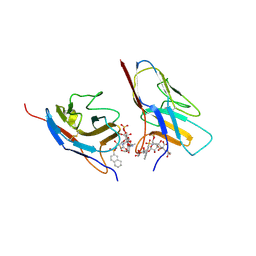 | | Crystal structure of the N-terminal domain of Siglec-8 in complex with sulfonamide sialoside analogue | | Descriptor: | (2~{S},4~{S},5~{R},6~{R})-5-acetamido-2-[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-acetamido-2-(hydroxymethyl)-4,6-bis(oxidanyl)oxan-3-yl]oxy-3,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-4-yl]oxy-6-[(1~{R},2~{R})-3-(naphthalen-2-ylsulfonylamino)-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Oyenarte, I, Atxabal, U, Nycholat, C, Franconetti, A, Quintana, J.I, Delgado, S, Unione, L, Paulson, J, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
6QB1
 
 | | 5676 | | Descriptor: | LEU-GLY-GLN-GLN-GLN-PRO-ALA-PRO-PRO-GLN-GLN-PRO-TYR | | Authors: | Calvanese, L, D'Auria, G, Falcigno, F. | | Deposit date: | 2018-12-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural insights on P31-43, a gliadin peptide able to promote an innate but not an adaptive response in celiac disease.
J.Pept.Sci., 25, 2019
|
|
6QB0
 
 | | 5675 | | Descriptor: | LEU-GLY-GLN-GLN-GLN-ALA-PHE-PRO-PRO-GLN-GLN-PRO-TYR | | Authors: | Calvanese, L, D'Auria, G, Falcigno, F. | | Deposit date: | 2018-12-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural insights on P31-43, a gliadin peptide able to promote an innate but not an adaptive response in celiac disease.
J.Pept.Sci., 25, 2019
|
|
6KBO
 
 | | Three-dimensional LPS bound structure of VG16KRKP-KYE28. | | Descriptor: | Heparin cofactor 2, VG16KRKP | | Authors: | Ilyas, H, Bhunia, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism.
J.Biol.Chem., 294, 2019
|
|
6K4I
 
 | | The partially disordered conformation of ubiquitin (Q41N variant) | | Descriptor: | ubiquitin | | Authors: | Wakamoto, T, Ikeya, T, Kitazawa, S, Baxter, N.J, Williamson, M.P, Kitahara, R. | | Deposit date: | 2019-05-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic relaxation enhancement-assisted structural characterization of a partially disordered conformation of ubiquitin.
Protein Sci., 28, 2019
|
|
7MZZ
 
 | |
7MZW
 
 | |
7MZX
 
 | |
9BB1
 
 | |
9BB2
 
 | |
9BB6
 
 | |
9BB4
 
 | |
9BB3
 
 | |
9MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18N | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
3IR2
 
 | |
5GHB
 
 | |
5GHC
 
 | |
5GHD
 
 | |
